Ipomoea batatas, commonly known as sweet potato, is a nutritious root vegetable that belongs to the Convolvulaceae family. It is cultivated worldwide for its edible tuberous roots, which come in a variety of colors and are rich in carbohydrates, dietary fiber, vitamins, and minerals. Sweet potatoes are not only a staple food source but also offer potential health benefits due to their bioactive compounds.
Metabolomics, a branch of omics sciences, focuses on comprehensively studying the small molecules (metabolites) present in biological systems. In the context of Ipomoea batatas, metabolomics analysis offers insights into the diverse metabolites that contribute to its nutritional value, flavor, color, and bioactivity.
By applying metabolomics techniques to Ipomoea batatas, researchers can gain a deeper understanding of its metabolome—capturing the snapshot of all metabolites present under specific conditions. This analysis helps uncover metabolic pathways, identify potential biomarkers, and explore how various factors like cultivation practices, environmental conditions, and genetic variations influence the plant's chemical composition.
At Creative Proteomics, we utilize advanced metabolomics technologies to unravel the complexity of Ipomoea batatas metabolism. Our targeted and untargeted approaches provide valuable insights into its nutritional content, bioactive compounds, and potential health benefits. Through our services, we contribute to the comprehensive exploration of sweet potato's metabolome, enhancing our understanding of its role in nutrition and human health.
Specific Ipomoea batatas Analysis Projects We Provide
Sweet Potato Metabolite Profiling: Comprehensive exploration of sweet potato metabolites to understand its nutritional value, color, flavor, and potential health benefits.
Bioactive Compounds Identification: Identification of bioactive compounds with potential health-promoting properties using untargeted metabolomics.
Flavor and Aroma Analysis: Profiling of volatile compounds influencing sweet potato flavor and aroma to enhance sensory attributes.
Varietal Differences Exploration: Comparison of metabolite profiles among sweet potato varieties to understand varietal differences in flavor, color, and bioactive compounds.
Quality Control in Sweet Potato Products: Assurance of product quality and authenticity through metabolomics analysis, reflecting processing methods and origin.
Impact of Processing Techniques: Analysis of processing effects on sweet potato metabolite composition and final product quality.
Environmental Stress Responses: Investigation of how sweet potato responds metabolically to environmental stresses such as drought and temperature fluctuations.
Sweet Potato Storage and Post-Harvest Research: Study of metabolite changes during storage and post-harvest processing, guiding preservation and quality enhancement.
Sweet Potato Salt and Cold Tolerance Research: Exploration of metabolic adaptations in response to salt and cold stress for breeding resilient varieties.
Nitrogen Metabolism Research: Analysis of nitrogen-related metabolites in sweet potatoes to enhance understanding of nitrogen utilization.
Biochemical Pathway Mapping: Elucidation of metabolic pathways leading to the synthesis of compounds like starches and antioxidants.
Integration with Genomics: Combination of metabolomics and genomic data to uncover the genetic basis of metabolite variation in sweet potatoes.
Chemical Composition and Quality Relationship Study: Investigation of the relationship between chemical composition and quality attributes of sweet potatoes.
Metabolic Engineering Research: Use of metabolic engineering to manipulate sweet potato metabolism and enhance desired compound accumulation.
Metabolism and Stress Adaptation Relationship Study: Exploration of sweet potato metabolism under stress conditions to uncover adaptive mechanisms.
Natural Medicinal Compounds Analysis: Identification of medicinal compounds in sweet potatoes, supporting traditional medicine and pharmacology studies.
Ipomoea batatas Metabolomics Analysis Techniques
Liquid Chromatography-Mass Spectrometry (LC-MS):
Liquid Chromatography-Mass Spectrometry (LC-MS) combines liquid chromatography separation with mass spectrometry detection. This versatile technique is employed to profile a wide range of metabolites in sweet potatoes, including both polar and non-polar compounds. At Creative Proteomics, we employ advanced LC-MS setups such as the Agilent 1290 Infinity LC System coupled with the Agilent 6550 iFunnel Q-TOF MS for accurate and comprehensive analysis.
Gas Chromatography-Mass Spectrometry (GC-MS):
Gas Chromatography-Mass Spectrometry (GC-MS) is specifically designed for analyzing volatile compounds, making it suitable for detecting aroma compounds in sweet potatoes. By separating compounds based on their volatility, GC-MS provides high-resolution spectra. Our expert team utilizes cutting-edge instrumentation like the Agilent 7890B GC System coupled with the Agilent 5977B MSD to precisely capture volatile profiles.
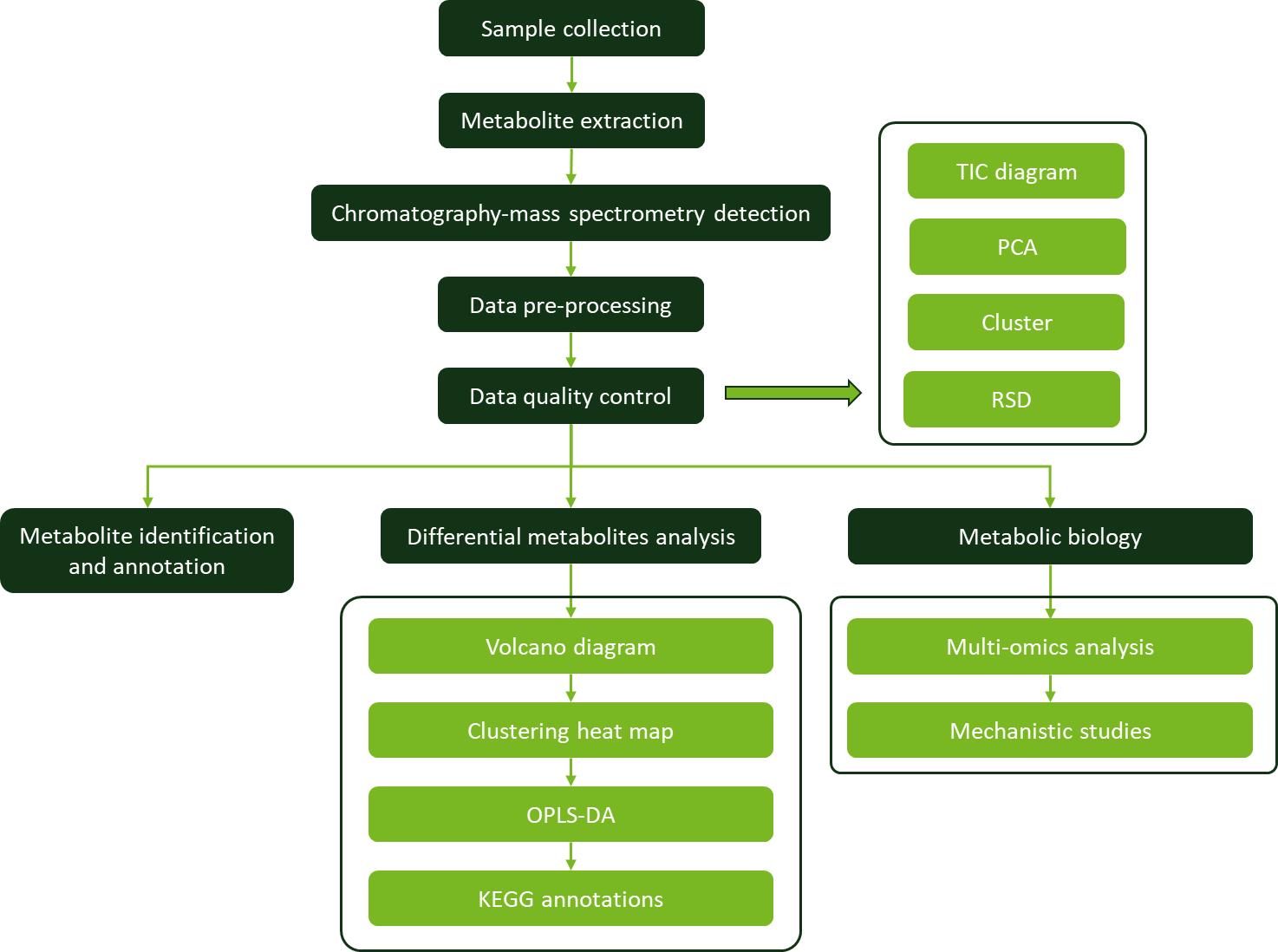 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
Sample Requirements for Ipomoea batatas Metabolomics
| Sample Type |
Sample Amount |
| Fresh Roots |
50-100 g |
| Dried Roots |
5-10 g |
| Root Extracts |
100-500 mg |
| Fresh Leaves |
50-100 g |
| Dried Leaves |
5-10 g |
| Leaf Extracts |
100-500 mg |
| Stems |
Depending on type |
| Stem Extracts |
100-500 mg |
| Flowers |
Depending on type |
| Flower Extracts |
100-500 mg |
| Seeds |
Depending on type |
| Seed Extracts |
100-500 mg |
| Tubers |
50-100 g |
| Tuber Extracts |
100-500 mg |
Case 1. Comparative Proteomic and Metabolomic Analysis of Two Sweetpotato Ecotypes Reveals Varied Nutrient Accumulation and Stress Response
Background:
Understanding the molecular basis of nutrient accumulation and stress responses in different sweetpotato ecotypes is crucial for enhancing crop productivity and nutritional quality. Varied nutrient allocation and stress adaptation are governed by complex interactions between proteins and metabolites within storage organs.
Samples:
The study focused on two contrasting ecotypes of sweetpotato: cv. OFSP and cv. WFSP. These ecotypes exhibit distinct responses to nutrient acquisition and environmental cues, making them suitable models to explore varietal differences in nutrient accumulation.
Methods:
Proteomic Analysis:
Protein Extraction: Tuberous root tissues were homogenized, and proteins were extracted using a suitable buffer. The extracted proteins were quantified, and their quality was assessed.
Protein Digestion: The extracted proteins were subjected to enzymatic digestion using trypsin, which cleaves proteins into smaller peptides. This digestion step is crucial for mass spectrometry-based analysis.
LC-MS/MS Analysis: The digested peptides were analyzed using Liquid Chromatography-Mass Spectrometry (LC-MS/MS). This involves separating peptides based on their chemical properties using liquid chromatography and then subjecting them to mass spectrometry for identification.
Database Search: The acquired mass spectrometry data were analyzed using multiple search engines. This approach increased the robustness of protein identification. The identified peptides were matched to protein sequences in databases.
Functional Annotation: The identified proteins were functionally annotated to understand their subcellular localization, pathway affiliations, and physicochemical characteristics. This helped categorize the proteins based on their roles in different cellular processes.
Metabolomic Analysis:
Metabolite Extraction: Tuberous root samples were subjected to metabolite extraction using appropriate solvents. This extraction process aimed to capture a wide range of metabolites present in the samples.
Metabolite Profiling: The extracted metabolites were profiled using various analytical techniques, such as mass spectrometry and chromatography. This allowed the identification and quantification of different metabolites present in the samples.
Metabolite Classification: The identified metabolites were categorized into different groups based on their molecular structures and putative functions. Common classes included organic acids, sugars, amino acids, sugar alcohols, and other miscellaneous compounds.
Comparison and Analysis: The metabolite profiles of the two ecotypes were compared to identify common metabolites and ecotype-specific metabolites. This analysis provided insights into the diversity of metabolites in different sweetpotato ecotypes.
Data Integration and Interpretation:
The results of the proteomic and metabolomic analyses were integrated to gain a comprehensive understanding of the molecular basis of nutrient accumulation and stress responses in cv. OFSP and cv. WFSP. The overlapping and distinct components in terms of proteins and metabolites were analyzed in the context of their roles in various metabolic pathways and functions. This integration facilitated the interpretation of how these components interact and contribute to varietal differences in nutrient allocation and stress adaptation.
Results
The proteomic analysis led to the identification of 1541 and 1201 proteins in cv. OFSP and cv. WFSP, respectively. A diversity of proteins associated with carbohydrate metabolism, storage activities, protein metabolism, transcription, translation, and cell defense was observed in both ecotypes. The metabolome profiling identified 148 metabolites in cv. OFSP and 126 metabolites in cv. WFSP, showcasing the diversity of organic acids, sugars, amino acids, sugar alcohols, and other compounds in both ecotypes. Several metabolites were conserved between the ecotypes, while others showed ecotype-specific accumulation.
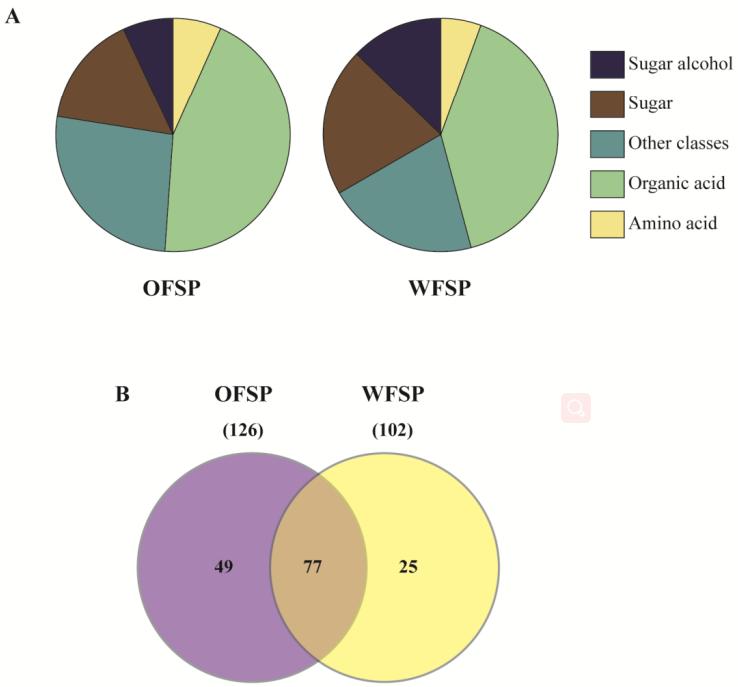 Comparative metabolome and distribution of metabolites.
Comparative metabolome and distribution of metabolites.
Reference
- Shekhar, Shubhendu, et al. "Comparison of proteomic and metabolomic profiles of two contrasting ecotypes of sweetpotato (Ipomoea batata L.)." Journal of Proteomics 143 (2016): 306-317.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service Comparative metabolome and distribution of metabolites.
Comparative metabolome and distribution of metabolites.