How to Ensure Quality Control in DIA Proteomics: Key Metrics & Checkpoints
Data-Independent Acquisition (DIA) proteomics has become a cornerstone for large-scale quantitative proteomics, enabling deep proteome coverage with exceptional reproducibility. However, the complexity and data intensity of DIA demand stringent quality control (QC) at every stage. Without robust QC, issues such as inconsistent retention times, inaccurate quantitation, or elevated false discovery rates (FDR) can compromise the integrity of biological conclusions.
At Creative Proteomics, we recognize that researchers need more than just raw data—they need confidence that every figure, every peptide ratio, and every pathway interpretation is based on rigorously validated results. That confidence can only be achieved through a robust and transparent quality control (QC) framework integrated throughout the DIA pipeline.
In this article, we will walk you through the metrics, checkpoints, and advanced strategies we employ to guarantee that each DIA proteomics project we deliver meets the highest scientific and publication standards.
Our Quality Control Philosophy in DIA Proteomics
In DIA proteomics, quality control is not a single step—it is the foundation upon which the entire workflow stands. Many service providers limit QC to final data verification, but this approach is insufficient for a method as data-intensive and sensitive as DIA. Our philosophy at Creative Proteomics is fundamentally different: we integrate QC checkpoints at every stage, ensuring issues are detected and corrected before they can compromise data integrity.
Core Principles Behind Our QC System
- Proactive, Not Reactive
QC is embedded as a continuous monitoring process, from sample receipt to data reporting, rather than being applied post-analysis. This means that any deviation—whether in LC pressure stability or mass accuracy—triggers immediate corrective measures, minimizing the risk of compromised datasets.
- Multi-Layered Validation
We combine hardware-level QC, such as monitoring vacuum system integrity and ion transmission efficiency, with software-based validation, including retention time alignment checks, precursor/fragment correlation scoring, and global FDR estimation. This dual approach ensures both physical performance and computational accuracy.
- Compliance and Transparency
Our QC thresholds align with international proteomics standards (e.g., HUPO, PSI), maintaining ≤1% protein-level FDR and ≤15% technical CV. For every project, we provide a full QC report, including system suitability results, performance logs, and visual summaries, enabling clients to confidently reference data quality in publications or audits.
Choosing a QC-driven provider is not about avoiding errors; it's about guaranteeing reproducibility, publication readiness, and regulatory alignment. Our philosophy reduces project risk, enhances cross-batch comparability, and ultimately accelerates scientific insights without costly repeat experiments.
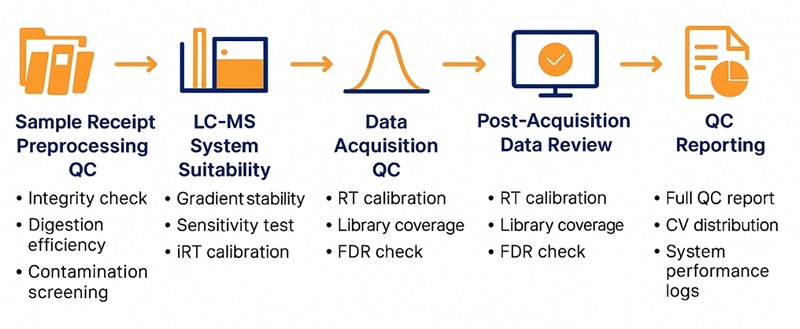 DIA QC Workflow Diagram: Five key checkpoints from sample receipt to QC reporting to ensure data stability and reproducibility.
DIA QC Workflow Diagram: Five key checkpoints from sample receipt to QC reporting to ensure data stability and reproducibility.
Key QC Metrics We Monitor
The robustness of DIA proteomics depends on systematic monitoring of critical performance indicators. Unlike targeted assays or DDA workflows, DIA captures all precursors across m/z ranges, generating complex datasets where even minor variability can propagate into significant biases. At Creative Proteomics, we have defined a rigorous set of quantifiable QC metrics covering instrument performance, chromatography stability, identification confidence, and quantitative reproducibility.
Mass Spectrometer Performance
Accurate mass measurement is essential for matching fragment ions to spectral libraries. Even slight mass errors lead to misidentification and increased false discovery rates (FDR).
Our Approach
- Mass Accuracy: Maintained within ±5 ppm throughout acquisition, validated using calibration peptides.
- Resolution & Sensitivity: Routinely verified with reference standards to ensure stable detection of low-abundance peptides.
- Signal-to-Noise Ratio (S/N): Tracked to detect early ion source contamination or detector aging.
These controls guarantee reliable protein IDs, reduce missing values, and safeguard the interpretability of downstream biological analysis.
Chromatographic Stability
DIA workflows rely on precise retention time (RT) alignment for accurate peptide quantitation and data normalization. RT drift can distort feature matching and impair reproducibility.
Our Approach
- RT Drift Threshold: Maintained under 0.5 minutes between runs, verified using iRT peptide standards.
- Peak Symmetry and Width: Monitored to detect LC column degradation and prevent compromised separation efficiency.
Ensures consistent peptide elution profiles, supporting robust cross-sample comparisons in multi-batch studies.
Quantitative Reproducibility
In biomarker discovery and comparative studies, quantitative consistency is paramount. Technical variation can overshadow true biological differences, leading to false conclusions.
Our Approach
- Coefficient of Variation (CV): For technical replicates, CV ≤15% across the majority of quantified proteins.
- Missing Value Rate: Kept below 20% through optimized sample prep and acquisition settings.
Identification Confidence
Comprehensive DIA datasets must balance depth of coverage with identification accuracy. Uncontrolled FDR inflates protein lists with false positives, reducing biological relevance.
Our Approach
- FDR Control: Strict thresholds of ≤1% applied at both PSM and protein levels.
- Spectral Match Quality: Verified through advanced scoring algorithms in Spectronaut/DIA-NN, leveraging fragment intensity correlations.
QC Metrics Summary Table
| QC Parameter | Acceptance Criteria | Why It Matters |
| Mass Accuracy | ±5 ppm | Prevents misidentification |
| RT Drift | <0.5 min | Ensures alignment for quantitation |
| Technical CV | ≤15% | Guarantees reproducibility |
| Protein-Level FDR | ≤1% | Validates identification confidence |
| Missing Value Rate | <20% | Maintains dataset completeness |
Critical Checkpoints in Our DIA Workflow
Sample Receipt & Pre-Processing QC
Poor sample integrity or suboptimal digestion efficiency introduces variability that no downstream correction can resolve.
Our Approach
- Integrity Verification: Assess protein integrity using SDS-PAGE and total protein quantification before digestion.
- Digestion Efficiency Check: Evaluate missed cleavage rates by preliminary MS scans, ensuring peptide representation is uniform.
- Contamination Screening: Monitor for keratin and other common contaminants to avoid false identifications.
LC-MS System Suitability
Stable chromatography and calibrated MS performance are prerequisites for consistent data acquisition across runs and projects.
Our Approach
- iRT Standards: Added to every batch to validate retention time consistency and LC gradient integrity.
- Performance Test Mix: Used to monitor MS sensitivity, resolution, and ion transmission efficiency before production runs.
Client Benefit: Prevents batch-to-batch drift and ensures your multi-condition comparisons remain scientifically valid.
Data Acquisition QC
DIA relies on systematic coverage of all precursors; any misconfiguration in isolation windows or acquisition cycles can cause significant data loss.
Our Approach
- Real-Time Monitoring: Automated checks for ion accumulation time, cycle completeness, and signal stability during acquisition.
- Window Scheme Validation: Confirm that the chosen m/z isolation strategy optimally balances coverage and selectivity for your proteome.
Post-Acquisition Data Review
QC does not stop when data collection ends; misaligned retention times or insufficient library coverage can still derail quantitation accuracy.
Our Approach
- RT Calibration Check: Use internal standards to confirm retention time alignment across runs.
- Library Coverage Verification: Assess whether reference libraries sufficiently represent target proteomes.
QC Reporting and Transparency
Clients need proof of quality, not just assurances.
Our Approach
- Comprehensive QC Reports: Delivered with every project, including key plots such as CV distributions, correlation heatmaps, PCA for batch effect analysis, and system performance logs.
- Full Traceability: All QC actions are documented, allowing data audits and regulatory compliance.
Advanced QC Strategies That Differentiate Our Service
- Isotope-Labeled Standards
We incorporate stable isotope-labeled peptides as internal references across runs, ensuring absolute quantitation accuracy and robust normalization. This mitigates variability introduced by sample preparation or instrument drift, resulting in consistently reliable measurements.
- Automated QC Tracking
Our workflow is fully integrated with leading DIA software platforms (Spectronaut, DIA-NN), enabling continuous monitoring of key parameters such as retention time alignment, mass accuracy, and signal stability. This automation minimizes human error and accelerates issue detection.
- Bridge Sample Design
For multi-batch or longitudinal studies, we implement bridge samples and pooled QC references, controlling batch effects and enabling statistically sound comparisons across extended timeframes.
- AI-Driven Anomaly Detection
By applying machine learning algorithms to large datasets, we identify outliers, systematic drifts, and unexpected technical variances before they impact downstream biological interpretation. This proactive approach ensures data consistency even in high-throughput and multi-lab collaborations.
Deliverables: What You Receive
- Full QC Report
A detailed summary of key quality metrics (e.g., CV, FDR, retention time stability), acceptance thresholds, and final performance evaluations. This document serves as auditable evidence of data quality for publications, funding reports, or compliance audits.
- Instrument Performance Logs
Complete logs of LC-MS performance during acquisition, including gradient stability, calibration status, and sensitivity validation results.
- Correlation Matrices & PCA Plots
Visual evidence of reproducibility and batch-effect control, allowing clients to quickly assess data integrity across conditions.
- Data Traceability
Access to full raw data files, along with version-controlled processing parameters, ensures complete transparency and supports re-analysis or independent validation if required.
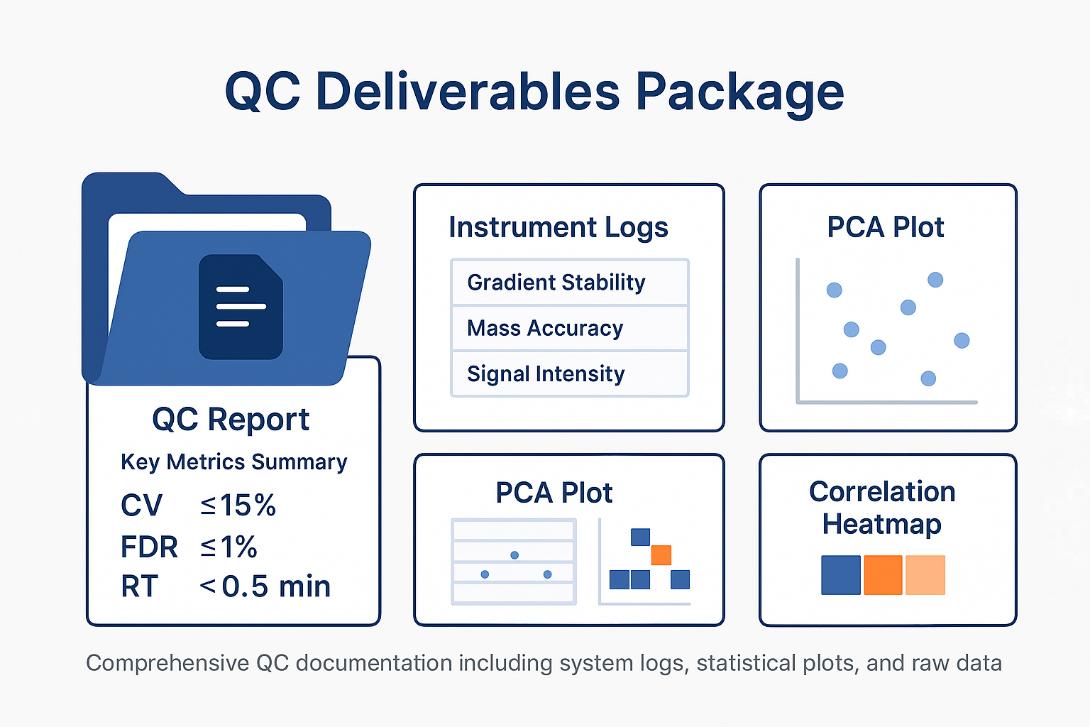 QC Deliverables Package: Includes QC report, instrument performance logs, PCA plot, and correlation heatmap to ensure full transparency and data reliability.
QC Deliverables Package: Includes QC report, instrument performance logs, PCA plot, and correlation heatmap to ensure full transparency and data reliability.
Ready to elevate your proteomics research?
Explore our DIA Proteomics Service
References:
- Zhang, Fangfei, et al. "Data‐independent acquisition mass spectrometry‐based proteomics and software tools: a glimpse in 2020." Proteomics 20.17-18 (2020): 1900276.
- Ludwig, Christian, et al. "Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial." Molecular Systems Biology 14.8 (2018): e8126.
- Bruderer, Roland, et al. "Analysis of 1508 plasma samples by capillary-flow data-independent acquisition profiles proteomics of weight loss and maintenance." Molecular & Cellular Proteomics 18.6 (2019): 1242-1254.
- Navarro, Pedro, et al. "A multicenter study benchmarks software tools for label-free proteome quantification." Nature Biotechnology 34.11 (2016): 1130-1136.
- Gillet, Ludovic C., et al. "Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis." Molecular & Cellular Proteomics 11.6 (2012): O111.016717. doi:10.1074/mcp.O111.016717.


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)