Why DIA Proteomics?
Unlock Reliable, Deep Proteome Insights with Data-Independent Acquisition
Traditional data-dependent acquisition (DDA) methods, while powerful, often struggle with missing values, limited reproducibility, and inconsistent quantification—especially when analyzing complex biological samples or large study cohorts. Researchers increasingly turn to data-independent acquisition (DIA) proteomics to overcome these limitations and achieve robust, high-confidence results.
Content Guide
- Advantages of DIA
- Discovery and Targeted Proteomics
- Workflow and Platform
- When Choose DIA
- Sample Requirements
- Deliverables
DIA vs DDA: Why Make the Switch?
Unlike DDA, where only the most intense precursor ions are selected for fragmentation in each cycle, data-independent acquisition mass spectrometry systematically fragments all precursor ions within defined m/z windows. This fundamental difference provides:
Consistent Quantification Across Runs
DIA ensures the same set of peptides is measured every time, reducing missing values and improving reproducibility in both discovery and targeted studies.
Enhanced Proteome Coverage
By capturing fragment ion data for virtually all detectable peptides, DIA proteomics enables deeper exploration of the proteome—even for low-abundance proteins often missed in DDA workflows.
High Throughput for Large Studies
DIA is ideal for large-scale experiments, such as biomarker discovery and clinical cohort studies, where consistent quantitative data across many samples is critical.
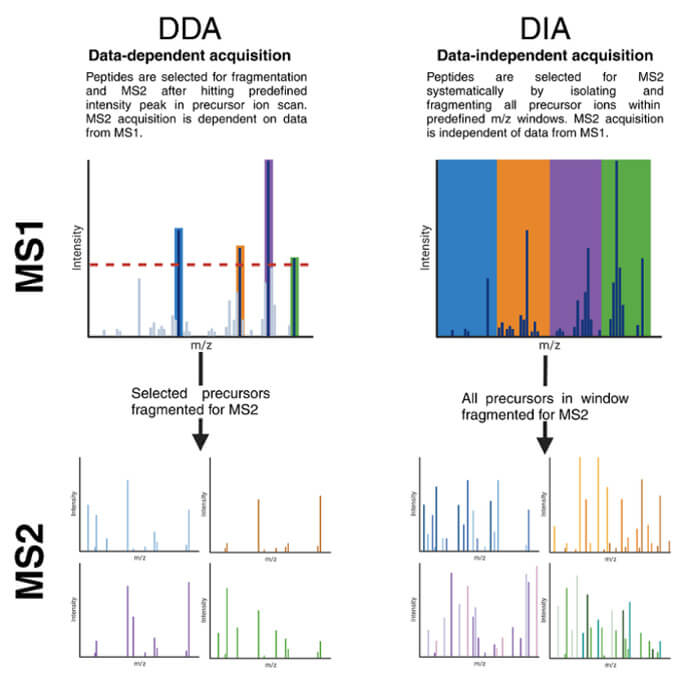 Difference between DDA and DIA. In DDA, precursors are selected for fragmentation and MS2 (Fig from Ward et al., 2024. Doi: 10.1101/2024.02.23.581160)
Difference between DDA and DIA. In DDA, precursors are selected for fragmentation and MS2 (Fig from Ward et al., 2024. Doi: 10.1101/2024.02.23.581160)
Advantages of Data-Independent Acquisition Proteomics
Deeper Proteome Coverage
8,000–10,000 Proteins / Run
Quantify up to 50% more proteins per run than DDA, enabling comprehensive proteome profiling even in complex biological samples.
Superior Reproducibility
< 10% CV Variability
Achieve consistent quantification across replicates, crucial for reliable statistical analysis and large-scale research projects.
Minimal Missing Data
< 2% Missing Values
Keep missing data rates below 2%, compared to up to 40% in DDA, ensuring robust statistical analysis and reliable biological conclusions.
Wide Dynamic Range
5–6 Orders of Magnitude
Detect both high- and low-abundance proteins in the same run, enabling insights across diverse biological pathways and molecular processes.
Scalable for Large Studies
Hundreds of Samples Analyzed Consistently
Maintain stable performance and data quality across large cohorts, supporting biomarker discovery, drug development, and multi-site studies.
High-Resolution Compatibility
< 1–2 ppm Mass Accuracy
Integrate seamlessly with Orbitrap and TOF instruments for precise mass measurements and confident protein identification.
Transforming Discovery and Targeted Proteomics
From Comprehensive Discovery to Precise Quantification
At Creative Proteomics, DIA proteomics is more than just a high-throughput technology—it's a versatile platform enabling both broad exploration and focused validation within the same workflow.
DIA Shotgun Proteomics – Comprehensive Discovery
- Unbiased Proteome Coverage
Capture a global snapshot of protein expression across diverse biological samples, including low-abundance proteins often missed in conventional approaches. - Efficient Hypothesis Generation
Identify novel protein candidates, pathways, and molecular interactions to support exploratory research and scientific discovery.
This approach is ideal for investigating biological mechanisms, characterizing complex systems, and generating data for future targeted studies.
DIA Targeted Proteomics – Focused Validation
- Retrospective Data Mining
Extract quantitative insights on specific proteins of interest from existing DIA datasets without needing additional analyses. - High Precision for Known Targets
Perform precise quantification of defined protein panels to support research validation, reproducibility studies, and hypothesis testing.
This targeted approach enhances confidence in results for exploratory and preclinical research, while keeping projects efficient and cost-effective.
Step-by-Step DIA Proteomics Workflow
Designed for Reliable, In-Depth Results
At Creative Proteomics, we tailor our data-independent acquisition proteomics workflow to answer your specific research questions. From discovery to targeted validation, we ensure consistent, high-quality data—every step of the way.
- Define study goals: discovery profiling, biomarker exploration, or targeted analysis
- Recommend suitable platforms and DIA strategies
- Matrix-specific extraction protocols for high recovery
- Rigorous peptide cleanup and quality checks
- Capture all peptides in each run, not just the top signals
- High-resolution instruments (Orbitrap/TOF) ensure <1–2 ppm mass accuracy
- Advanced software identifies and quantifies thousands of proteins
- Maintain <2% missing data, even across large cohorts
- Detect significant protein changes
- Map results into biological pathways for deeper insights
- Receive: Raw and processed data; Statistical analyses; Custom bioinformatics reports.
- Flexible formats to fit your downstream tools
- Library-free workflows → start projects even without existing spectral libraries.
- Low-input precision → high-quality results from minimal sample amounts.
- Fast turnaround → speed up your research timelines.
- Expert support → guidance from data analysis to next steps.
What Makes Our DIA Proteomics Service Stand Out?
Orbitrap-Based Systems
- Ultra-high mass accuracy (<1–2 ppm) and excellent resolution make Orbitrap instruments ideal for complex samples like tissue lysates, plasma, or serum.
- Particularly suited for projects demanding in-depth proteome coverage, confident peptide identification, and precise quantification.
- Frequently chosen for biomarker discovery studies where subtle changes in protein levels matter.

Time-of-Flight (TOF) Systems
- Known for fast acquisition speeds and high dynamic range, TOF instruments excel in large-scale studies requiring rapid throughput.
- Ideal for high-volume projects such as population-scale cohort studies, where hundreds of samples need to be processed consistently.
- Often selected for exploratory experiments where speed and scalability are priorities.
Trapped Ion Mobility Spectrometry (TIMS) – 4D DIA
- Adds a fourth dimension of separation based on ion mobility, effectively distinguishing co-eluting peptides and reducing signal interference.
- Improves selectivity and sensitivity, especially in complex matrices like plasma or tissue where co-elution is common.
- Particularly valuable for targeted studies focusing on low-abundance proteins or detailed pathway analyses.
When Should You Choose DIA Proteomics for Your Research?
Biomarker Discovery & Validation
- Detect low-abundance proteins for discovering novel disease biomarkers.
- Achieve consistent results suitable for multi-center studies and downstream validation.
Large-Scale Cohort
Studies
- Minimize missing data across hundreds of samples.
- Ideal for population health research, clinical trials, and epidemiological studies.
Drug Response and Mechanistic Studies
- Quantify protein changes in response to drug treatment or environmental stimuli.
- Useful for preclinical research and understanding drug mechanisms.
Pathway and Systems Biology
- Map comprehensive DIA datasets to biological pathways.
- Identify signaling networks and molecular interactions.
Discovery in Novel Organisms or Conditions
- Perform DIA without pre-existing spectral libraries.
- Suitable for non-model organisms, new disease models, or uncharacterized biological systems.
How to Prepare Samples for DIA Proteomics: Types, Amounts, and Tips
Proper sample preparation is essential for achieving accurate, reproducible results in DIA proteomics. Here's a detailed guide to help you plan your project effectively.

Storage and Shipping Tips
- Store all samples at –80°C unless otherwise noted.
- Avoid repeated freeze-thaw cycles.
- Ship samples on dry ice for long-distance transport.
- Include a detailed sample manifest with every shipment.
| Sample Type | Recommended Amount | Preferred Storage | Special Notes |
|---|---|---|---|
| Cell pellets | ≥ 1–5 million cells | –80°C | Lower amounts possible for targeted analyses. Avoid excessive detergent in lysis buffer. |
| Tissue lysates | ≥ 20–50 mg wet weight | –80°C | Homogenize thoroughly. Snap-freeze immediately after collection. |
| Plasma / Serum | ≥ 50–100 µL | –80°C | Collect using standard anticoagulants. Prefer EDTA over heparin. |
| Other biofluids | ≥ 100–200 µL | –80°C | Contact us for compatibility with urine, CSF, saliva, etc. |
| Exosomes | Contact us | –80°C | Requires specific isolation protocols. Discuss with us for guidance. |
| FFPE tissues | Consult us | Room temp / –20°C | Compatibility varies. Older FFPE blocks may yield lower-quality data. |
Not sure whether your samples meet the requirements?
Contact us — we're happy to help design the best strategy for your DIA proteomics study.
What You'll Receive from Our DIA Proteomics Service
At Creative Proteomics, we know that high-quality results are more than just numbers—they're the insights that drive your research forward. Here's what you'll receive when partnering with us for DIA proteomics:
Heatmap
Volcano Plot
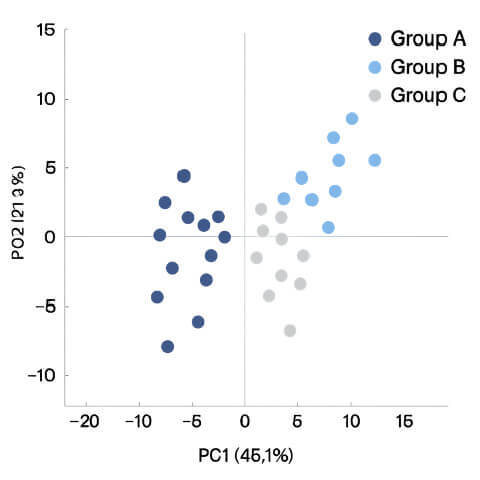
PCA Plot
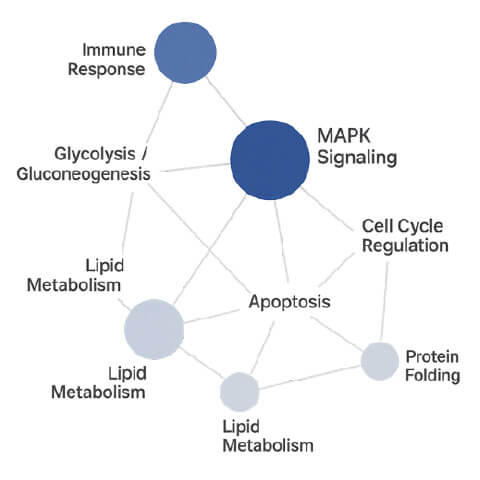
Pathway Network Diagram
Raw and Processed Data
- Access to raw mass spectrometry files for transparency and independent validation.
- Clean, organized quantification tables in user-friendly formats (.xlsx, .txt) for easy downstream analysis.
Comprehensive Quality Control Reports
- Detailed QC metrics covering instrument performance, calibration status, and run-to-run consistency.
- Visual plots including Total Ion Chromatograms (TICs) and peak count charts to verify data quality.
Differential Expression Analysis (Optional)
- Tables listing significantly changing proteins across experimental groups.
- Statistical outputs including fold changes, p-values, and FDR to support your conclusions.
Biological Interpretation & Pathway Analysis (Optional)
- Enrichment analyses highlighting key biological pathways and processes impacted in your study.
- Visual diagrams and pathway maps for biological context and deeper insights.
Publication-Ready Visualizations (Optional)
Included in your deliverables are high-quality, ready-to-use graphics:
- Heatmap → Reveals expression patterns across samples.
- Volcano Plot → Highlights significantly regulated proteins at a glance.
- PCA Plot → Visualizes how samples cluster or separate.
- Pathway Network Diagram → Maps biological pathways and interactions.
Yes. DIA proteomics data can be integrated with:
- Transcriptomics
- Metabolomics
- Phosphoproteomics
Reproducibility is a major strength of DIA. We ensure this by:
- Using consistent acquisition parameters across all runs.
- Incorporating internal standards.
- Performing rigorous QC checks on signal intensity and retention time shifts.
Yes. DIA data is complex and typically requires:
- Spectral deconvolution software
- Specialized search engines for library-free analysis
- Dedicated pipelines for quantification and FDR control
Yes—but with caveats. DIA can detect certain PTMs like phosphorylation or acetylation, but:
- Sensitivity for PTMs can be lower than for standard peptides.
- Enrichment steps (e.g., phosphopeptide enrichment) are often recommended.
4D-DIA uses ion mobility separation in addition to mass-to-charge and retention time. It:
- Separates co-eluting peptides
- Improves sensitivity
- Enhances identification in complex matrices like plasma or tissues
Yes. Our workflows can work with as little as:
- 50–100 ng peptides for certain discovery studies
- Lower inputs for targeted assays
Yes. We often help clients:
- Plan biological replicates
- Choose controls and experimental groups
- Decide on fractionation strategies
- Estimate sample size needed for statistical power
Success Stories with DIA Proteomics
Large-Scale DIA Proteomics in Mouse Development
Journal: Nature Communications · Published: 2024
Study Scope
Researchers performed the first comprehensive multi-organ proteome atlas covering mouse development from infancy to adulthood. The study analyzed:
- 10 organs: brain, heart, lung, liver, kidney, spleen, stomach, intestine, muscle, skin
- 3 developmental stages: 1 week, 4 weeks, 8 weeks
- Both sexes included (male and female)
- Total samples: 300 (10 organs × 3 time points × 2 sexes × 5 replicates)
This is one of the largest DIA-based proteomics datasets for developmental biology.
Proteomic Coverage and Data Quality
- Detected and quantified 11,533 protein groups across all organs
- Achieved dynamic range spanning ~10 orders of magnitude
- Median correlation coefficient between biological replicates reached 0.974, indicating excellent reproducibility
- Coefficient of variation (CV) < 30% in most samples
- No significant batch effects detected in PCA analysis
This confirms the robustness and stability of DIA workflows even in large-scale projects.
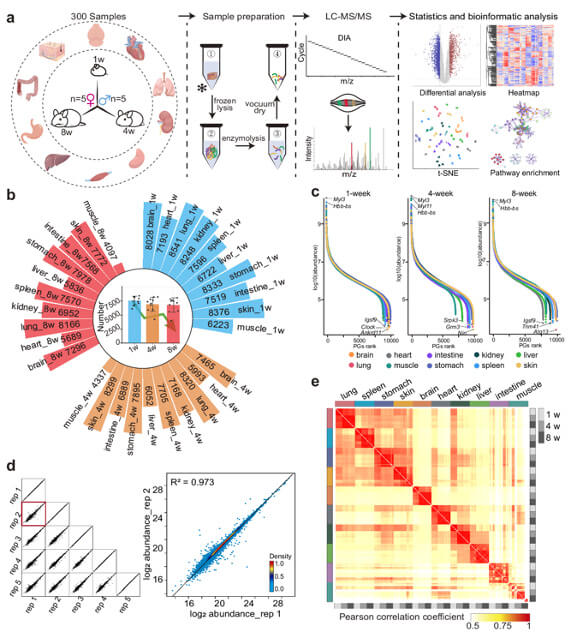 Proteome atlas of ten mouse organs from infancy to adulthood.
Proteome atlas of ten mouse organs from infancy to adulthood.
Biological Insights
The study revealed:
- 115 age-related proteins co-expressed across all organs, indicating conserved developmental regulators
- Key pathways affected include: Spliceosome regulation; Oxidative phosphorylation; Insulin signaling
- Organ-specific expression patterns and sexual dimorphism, notably in the liver
- Dynamic changes in pathways like fatty acid metabolism and cell cycle regulation
- Detection of unique developmental protein signatures specific to each organ and time point
These findings demonstrate DIA's capability to capture both global and nuanced biological changes.
Technical Highlights
- DIA data acquired on Q Exactive HF-X Orbitrap and timsTOF flex platforms
- Employed DIA-NN deep learning algorithms for library-free analysis:
- Improved detection of low-abundance peptides
- Reduced missing values
- Quality control included:
- Internal standards (iRT peptides)
- Hela digest controls for instrument performance
- Randomized measurement orders to test for batch effects
This setup exemplifies state-of-the-art DIA workflows suitable for large-scale, high-complexity studies.
Why It Matters
This study is a compelling example of how DIA proteomics can:
- Handle large sample numbers without compromising data quality
- Reveal intricate biological processes across multiple tissues
- Provide reproducible and quantitative data ideal for systems biology, biomarker discovery, and developmental studies
Reference
Wang, Qingwen, et al. "The mouse multi-organ proteome from infancy to adulthood." Nature Communications 15.1 (2024): 5752.



