4D-Acetylation Proteomics Services
Protein acetylation modifications are regulated by acetyltransferases and deacetylases and vary with metabolic environments. Their modifications often occur on metabolic enzymes, which regulate metabolic pathways and metabolic enzyme activities and participate in downstream metabolite changes. Protein acetylation is the most common type of acylation modification and refers to covalent binding acetyl groups (e.g., acetyl coenzyme A and other donors) to lysine residues of substrate proteins catalyzed by acetyltransferases. The above process mainly occurs at the theε-NH2 position of the protein lysine residue, and its molecular weight will increase by 42.01Da. Furthermore, mass spectrometry can accurately determine whether the molecular weight has a corresponding mass shift and analyze the acylated modified peptides and sites.
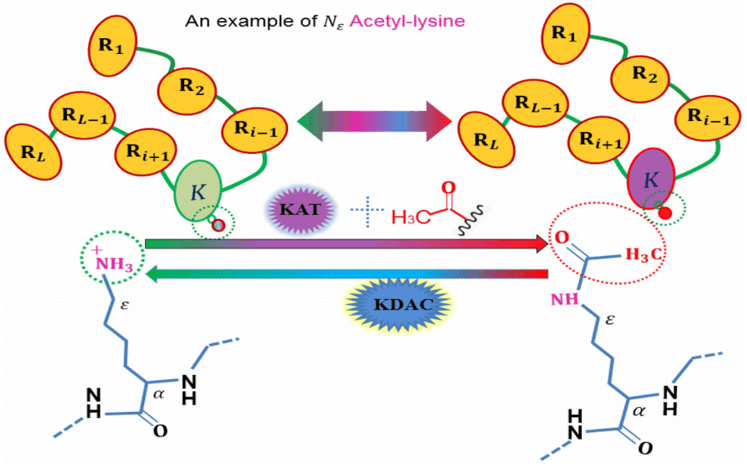 An illustration to show the acetylation protein. (Wangren Qiu et al. 2019)
An illustration to show the acetylation protein. (Wangren Qiu et al. 2019)
Our 4D-phosphoproteomics Services
Creative Proteomics provides 4D-label free acetylation proteomics, using the ion flow separation, which can effectively distinguish modified tautomeric peptides according to the shape and cross-sectional area properties of the molecule. This service improves the accuracy and depth of identification of protein acetylation modifications and is more suitable for the analysis of micro samples.
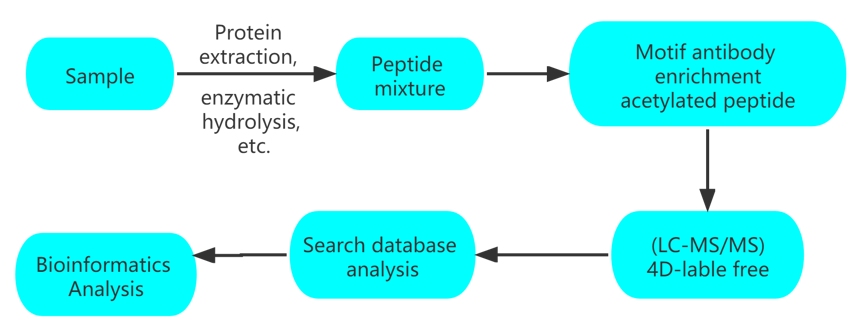
Workflow
| Type | Modification site | Enrichment method | Suggestion |
| 4D-label free acetylation | Lys | Acetyl-Lysine motif antibody | 1) Micro sample analysis |
| 4D-label free Succinylation | Lys | Succinyl-Lysine motif antibody | 2) Cost-effective analysis |
| 4D-label free Propionylation | Lys | Propionyl-Lysine motif antibody | 3) Experimental design with large differences in expression patterns between samples |
| 4D-label free Malonylation | Lys | Malonyl-Lysine motif antibody | |
| 4D-label free Glutarylation | Lys | Glutaryl-Lysine motif antibody |
Applications
- Regulation of energy metabolism.
- Metabolic syndrome research.
- Fungal-bacterial studies.
Data Analysis
| Standard Data Analysis Content | |
| Mass Spectrometry Data Analysis | Spectral peptide quality deviation distribution, peptide length distribution, unique peptide number distribution, protein coverage distribution |
| Protein Expression Analysis | Protein abundance value distribution of PTMs, protein abundance ratio distribution of PTMs between samples, PCA analysis, statistical analysis of significant differences |
| Protein Functional Analysis | Total protein and differential protein GO secondary classification, COG function classification, KEGG annotation, subcellular organelle location, domain annotation, signal peptide prediction and PPI prediction; differential protein GO, KEGG, domain enrichment analysis |
| Advanced Data Analysis Content | |
| Protein Gene Chromosome Localization | Obtain the distribution of genes encoding proteins on chromosomes |
| WGCNA Analysis | Predict functional clustering and network interactions by protein expression level; correlate with phenotype data to obtain key proteins or protein complexes that influence phenotype |
| Trend Cluster Analysis | Obtain protein expression trend patterns |
| Molecular Typing | Molecular typing for large cohort samples |
| Survival Curve Analysis | Study the relationship between influencing factors and survival time and outcome |
| ROC Curves | Evaluate predictive accuracy by combining specificity and sensitivity, such as biomarker impact assessment on tumor grade |
Advantages
Cutting-edge technology platform: new generation 4D platform with elevated identification depth and more accurate site location
Experimental data guarantee: gold standard antibodies, high-standard enrichment operations to ensure the stability of the experimental process
One-stop analysis: from PTMs omics to verification, upstream and downstream joint analysis
Huge research potential
Sample Requirements
| Sample Type | Protein | # of Cells | Animal Tissue | Plant Tissue | Blood | Urine | Serum | Microbes |
| Quantify | 100 ug | 1×107 cells | 1 g | 200 mg | 1 mL | 2 mL | 0.2-0.5 mL | Dry weighed: 200 mg |
Recommended Combination
Phenotype and mechanism study: acetylation + metabolomics.
References:
- Extreme Acetylation of the Cardiac Mitochondrial Proteome Does Not Promote Heart Failure. 2020.Circulation Research.
- Regulation of UCP1 and Mitochondrial Metabolism in Brown Adipose Tissue by Reversible Succinylation. 2019. Molecular Cell.


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)