DIA-MS Ubiquitinome Analysis Services
Ubiquitination modification is an important post-translational modification. The ubiquitin-proteasome system (UPS) mediates 80%-85% of protein degradation in eukaryotic organisms. In addition, ubiquitination modification can also directly affect the activity and localization of proteins, and regulate a variety of cellular activities including cell cycle, apoptosis, transcription regulation, DNA damage repair, and immune response. Thus, dysregulation of the UPS can lead to loss of cell cycle control and ultimately to cancer.
How to Study Ubiquitinome?
Although early studies of ubiquitinated proteins were performed on a target-by-target basis, mass spectrometry (MS)-based proteomics has facilitated the analysis of global ubiquitin signatures. The primary approach to ubiquitinome analysis relies on immunoaffinity purification and mass spectrometry-based detection of diethylene glycol-modified peptides (K-GG) that are generated by tryptic digestion of ubiquitin-modified proteins (referred to as ubiquitinomics or ubiquitinomics).
Thus far, a combination of label-free or isotope-based data-dependent acquisition (DDA) methods has been reported to quantify the ubiquitinome. Recently, data-independent acquisition (DIA) has emerged as a compelling alternative to DDA in proteomics analysis, enabling the identification of more proteins in different samples, more precise and accurate quantification of proteins with fewer missing values and higher identification rates over a larger dynamic range.
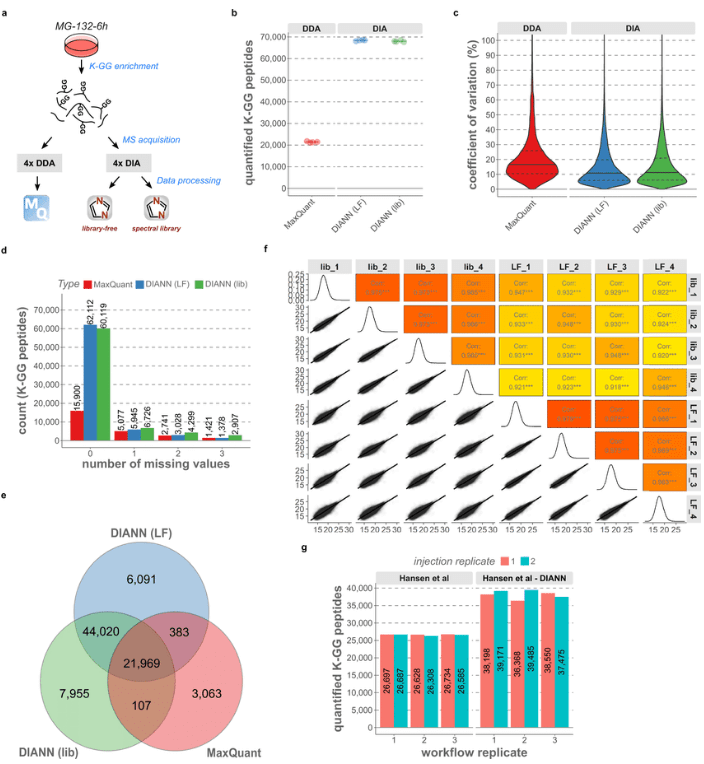 Side-by-side comparison between DDA and DIA for ubiquitinomics. (Martin Steger et, al. Nat Commun.2021)
Side-by-side comparison between DDA and DIA for ubiquitinomics. (Martin Steger et, al. Nat Commun.2021)
Our Large-Scale Ubiquitinome Profiling Services
Utilizing DIA MS technology, Creative Proteomics offers a large-scale ubiquitinated quantitative proteomic analysis service package. You just need to tell us your experimental purpose and send us your samples, we will take care of all the follow-ups of the project, including protein extraction, protein digestion, ubiquitinated peptide enrichment, peptide separation, mass spectrometry, mass spectrometry raw data analysis, and bioinformatics analysis.
We can detect ubiquitination on a large scale from complex biological samples on the discovery proteomics platform and absolute quantitative proteins for customized panels on a highly multiplexed targeted proteomics platform to provide a comprehensive overview of signaling events and pathway activity regulation.
Workflow
Our Ubiquitinome Profiling Services Include
- Enzyme digestion
- Plasma/serum protein depletion
- Peptide fractionation
- Peptide purification and concentration
- Quality control runs
- LC-MS/MS analysis
- Database search
Advantages
- Ubiquitinated peptide specific enrichment
- High sensitivity
- High accuracy
- High repetition rate
- High throughput
- Complete and comprehensive data analysis and presentation
Sample Requirements
1. Tissue Samples: Please send us the tissue samples on dry ice.
| Sample Type | Cell | Animal Tissue | Plant Tissue | Blood | Urine | Serum | Microbes |
| Quantify | 1×107 cells | 1 g | 200 mg | 1 mL | 2 mL | 0.2-0.5 mL | Dry weighed: 200 mg |
2. Protein Samples: Please make sure the total amount of protein is 2-5mg; ordinary tissue and cell lysate can be used for protein extraction.
3. Transportation: Please use sufficient amount of dry ice for transportation and try to use a faster postal delivery method to reduce the possibility of sample degradation during transportation.
4. Before the formal experiment, we will test the samples provided by you and start the formal experiment after passing the test.
Delivery
LC-MS/MS Report
References:
- Martin Steger et al. Time-resolved in vivo ubiquitinome profiling by DIA-MS reveals USP7 targets on a proteome-wide scale. Nat Commun.2021; 12: 5399.
- Fynn M. Hansen et al. Data-independent acquisition method for ubiquitinome analysis reveals regulation of circadian biology. Nat Commun.2021; 12: 254.


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)