4D-LFQ Metaproteomics Services
Meta-Omics is a subject that covers the study of metagenomics, metatranscriptomics and metaproteomics. In general, metagenomics research studies the species composition of microorganisms in the environment and has been carried out in full swing. Metaproteomics describes the protein expression of microorganisms and due to its complexity, it is still in its infancy. However, metaproteomics can analyze microbial communities' consistency, activity, and function at the protein level, which provides information that metagenomics cannot reveal.
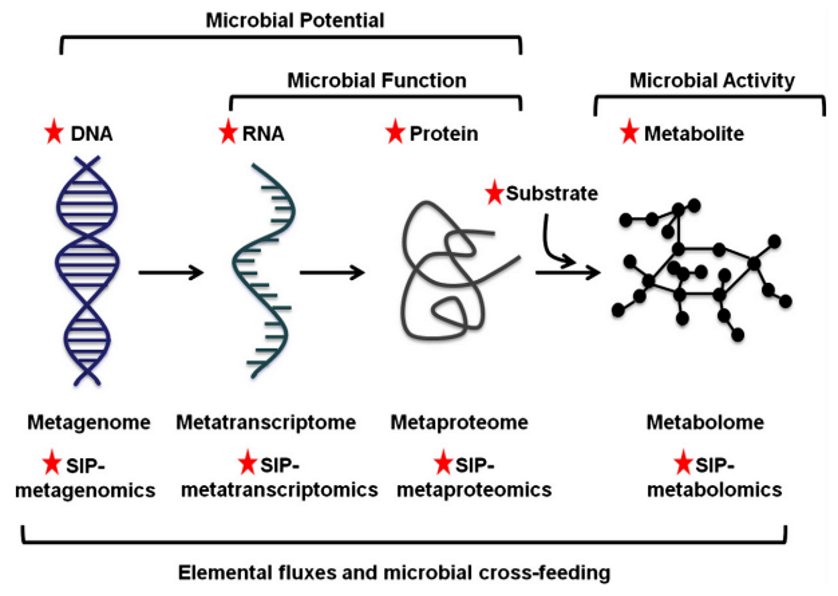 The multi-omics approach of microbiome[1]
The multi-omics approach of microbiome[1]
Metaproteomics uses mass spectrometry (MS)-based proteomics on studying all proteins of the microbial communities at a given moment in time. These communities can be part of environmental samples (soil, sludge, wastewater, ocean, etc.) or present in the host organism (human gut, mucosal cavity, etc.).
The research characteristics of macroproteomics include complex composition, various interferences; a wide variety of microorganisms; significant influence by external environmental factors (season, temperature, humidity, etc.); complex spatial location.
Difficulties of Mass Spectrometry Detection
The microbiome contains a wide variety of microorganisms and a vast abundance of protein
Strict mass spectrometry requirements, including scanning speed, resolution and quality accuracy
Specific commonly used quantitative proteomics techniques are not applicable
Our 4D-LFQ Metaproteomics Services
Using advanced high-resolution mass spectrometers Thermo Orbitrap, Creative Proteomics provides you with cutting-edge protein 4D-LFQ metaproteomics services for biomarker validation, clinical and industrial applications.
Workflow
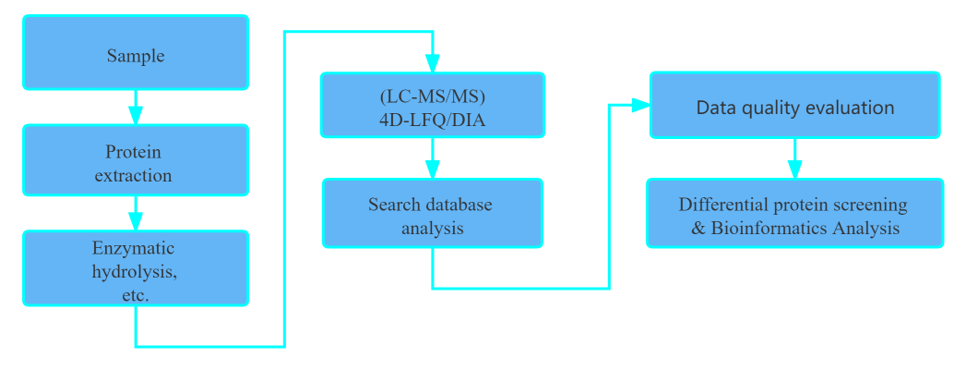
How to Perform Metaproteome Data Analysis?
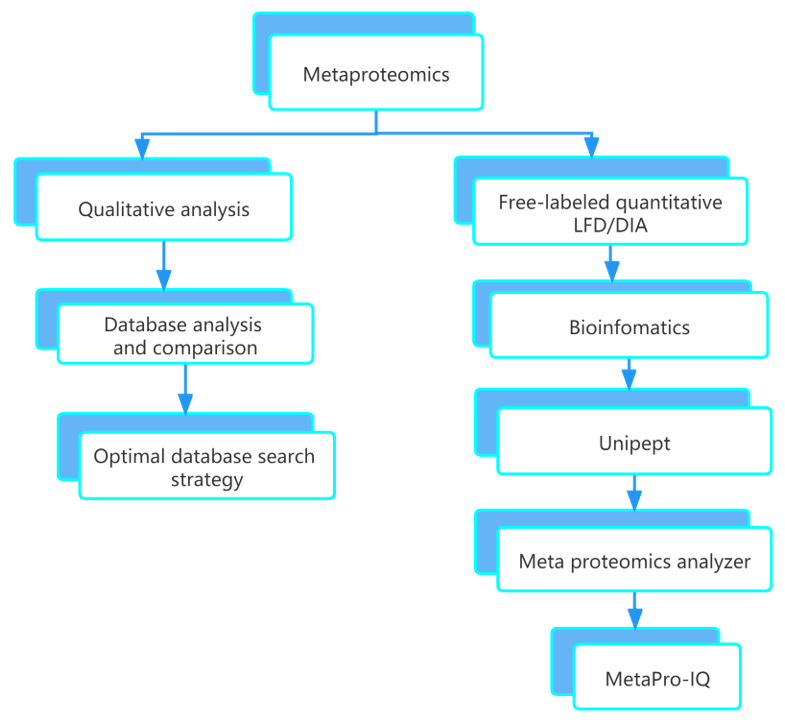
As a crucial feature for metaproteome analysis, its database is mainly obtained in two ways: 1) extracting the corresponding species sequence database from public database for analysis; 2) using the species sequence by metagenomic sequencing as reference sequence.
Features
Upregulated sensitivity: the minimum loading sample can be ng.
Advanced detection depth: detection depth increased by 20% in comparison to 3D instruments.
More accurate quantitative data: the fourth dimension of ion mobility separation provides more accurate quantitative data.
Higher stability and throughput: 4D-LFQ reduces the complexity of sample pre-processing and improves the stability of sizeable clinical cohort samples.
Applications
Gut flora analysis
Soil community analysis
Marine microbial analysis
Wastewater biological treatment
Plant root microenvironment
Fermented food microbial community analysis
Mucosal cavity analysis
etc.
Instrument
Tims TOF pro Thermo Orbitrap Fusion Thermo QE HF
Sample Requirements
Serum, plasma: 200 μL/sample
Cells, microorganisms: 1×107 cells/sample
Report
- Experimental steps
- Relevant experiment parameters
- Mass spectrometry spectra
- Raw data
- Proteomics analysis results
Reference:
- Florence Abram.Systems-based approaches to unravel multi-species microbial community functioning. Computational and Structural Biotechnology Journal 13 (2015) 24-32.


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)