Biomarker Proteomics Solutions
Biomarkers Development Pipeline
A biomarker is a characteristic biochemical indicator that can be objectively evaluated to determine the biological process of an organism. The examination of a disease-specific biomarker can play a key role in the diagnosis and prevention of the disease.
In the field of medical research, biomarkers are generally studied in three stages: discovery, verification, and validation. The screening of markers usually requires the use of high-throughput omics methods to conduct metabolomics or proteomics testing on large-scale clinical samples to screen for statistically significant differential metabolites or differential proteins, and then goes through a series of complex bioinformatics analysis to screen out target biomarkers.
In the next stage of verification, it is necessary to require large sample size validation of targeted proteomics or targeted metabolomics, statistical analysis, and target marker specificity and sensitivity testing for a smaller range of biomarkers. If more complete study results are desired, additional validation can be performed using clinical samples and combined with clinical data, such as ELISA and WB.
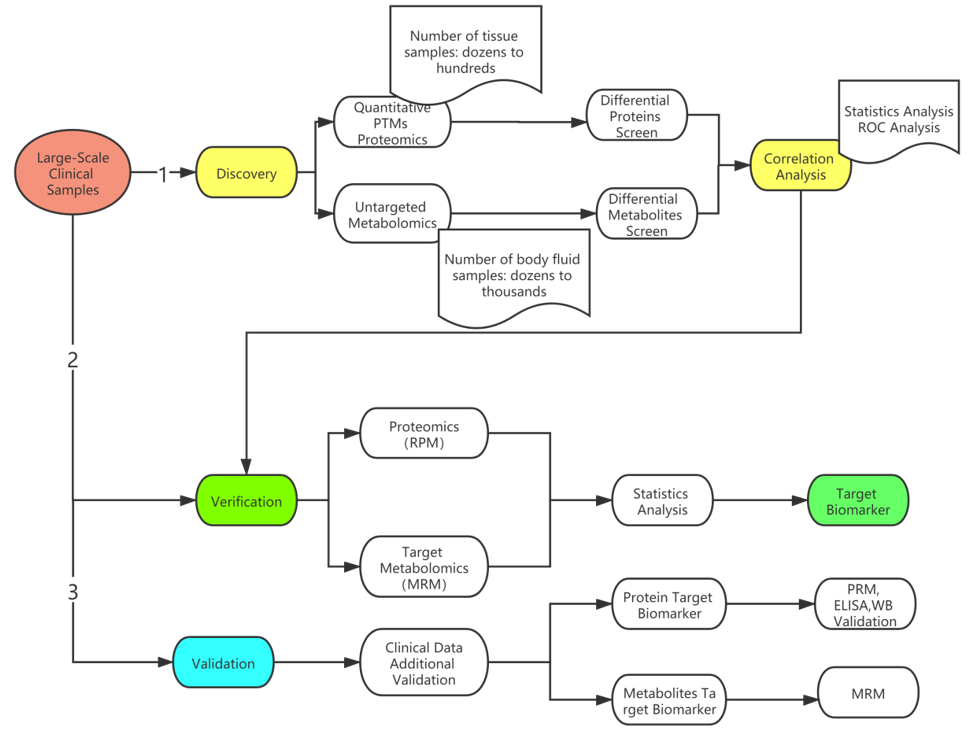 Biomarker research process
Biomarker research process
MS Technology for Biomarker Development
Quantitative mass spectrometry-based proteomics has become central to the first two steps in biomarker development pipeline as it is a powerful tool to identify and quantify novel candidate biomarkers for which abundance is altered in disease-specific conditions. Biomarker discovery and evaluation pipelines require robust and high-throughput protein assays to reduce attrition and deliver valuable candidate biomarkers at earlier stages.
Over the last decade, improvements in MS technology, particularly in acquisition speed, have made DDA (data-dependent acquisition) MS approach efficient for the rapid identification of a large number of peptides and proteins in complex mixtures. However, due to the stochastic nature of precursor ion selection, biased toward more abundant peptides, DDA often lacks reproducibility across samples and provides an incomplete peptide quantification matrix.
Thanks to the recent developments in high-resolution mass spectrometry instruments, data-independent acquisition (DIA) has become a realistic option to achieve the ultimate goal of proteomics: the comprehensive identification and quantification of proteins and peptides in complex biological matrices.
Our Biomarker Proteomics Services
Utilizing DIA-MS technology, Creative Proteomics offers a biomarker analysis service package. We can quantify and identify thousands of proteins per sample, and screen out significantly regulated proteins as biomarker candidates based on our discovey proteomics platform. These candidates can then be verified through our target proteomics platform, where up to 150 proteins per sample can be precisely quantified by relative quantification or absolute quantification of thousands of samples in a high-throughput mode.
All you need to do is to send us your samples and tell us your needs and we will take care of all the follow-ups of your project, including protein extraction, proteolysis, peptide separation, mass spectrometry analysis, mass spectrometry raw data analysis, and bioinformatics analysis.
Workflow
The depth and relatively straightforward nature of our workflow would make it a powerful new tool in the detection of biomarker.
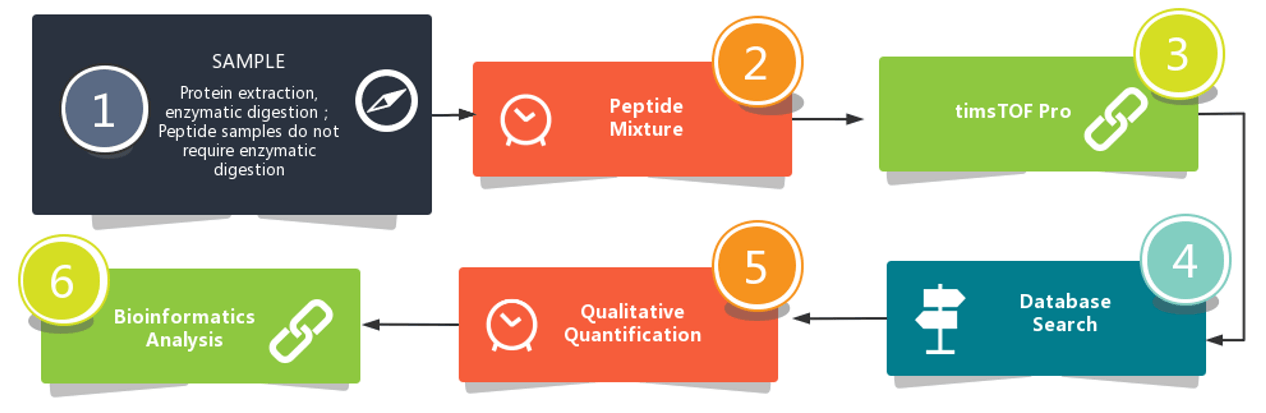
Bioinformatics Analysis
| Problems to Be Solved | Bioinformatics Analysis |
| Quality Assessment of Protein | SWATH Data Analysis |
| Protein Comparison of Different Samples | Multivariate PCA Analysis |
| Protein Statistical Analysis | Venn Diagram |
| Volcano Plot | |
| Functional Annotation | KEGG Annotation |
| GO Annotation | |
| COG Annotation | |
| Clustering Analysis | Hierarchical Clustering |
| K-Means Clustering | |
| Network Analysis | STRING Analysis |
Analytical Platforms
AB SCIEX Triple-TOF 5600-plus, Q-Exactive, Orbitrap Fusion
Benefits
- Early disease diagnosis
- Toxicity study
- Drug efficacy prediction
- Drug's mode of action analysis
Sample Requirements
| Sample Type | Protein | Cell | Animal Tissue | Plant Tissue | Blood | Urine | Serum | Microbes |
| Quantify | 100 ug | 1×107 cells | 1 g | 200 mg | 1 mL | 2 mL | 0.2-0.5 mL | Dry weighed: 200 mg |
Report
- Experimental steps
- Relevant experiment parameters
- Mass spectrometry pictures
- Raw data
- Proteomics analysis results
Since we started to provide DIA technology service, we have accumulated a lot of experience not only in the detection of a large cohort of conventional samples, but also in providing one-stop service of DIA+PRM+machine learning for biomarker studies, tumor-based proteomic typing, and large-scale clinical sample proteomics analysis based on next-generation chromatography (Evosep DIA). Free consultation is welcome.
Reference:
- Alexandra Kraut, Mathilde Louwagie, Christophe Bruley, Christophe Masselon, Yohann Coute', Virginie Brun, and Anne-Marie Hesse. Protein Biomarker Discovery in Non-depleted Serum by Spectral Library-Based Data-Independent Acquisition Mass Spectrometry. Proteomics for Biomarker Discovery. 2019.


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)