Proteomics Solutions for Genetic Association Study
Hundreds of genetic variants are associated with complex disorders, but the underlying molecular pathways often remain elusive. Genome-wide association studies (GWAS) are powerful approaches to identify genes responsible for diseases. GWAS has identified thousands of loci associated with complex traits, but pinpointing the causal genes in these loci and underlying molecular pathways is a daunting task. Because the genomic data information of GWAS contains coding, non-coding, functional and non-functional genetic variants, it is difficult to distinguish them in association studies.
In contrast, proteomic analysis of biological samples such as cells, tissues, and body fluids has yielded valuable insights into the complex processes of disease in recent years. Proteins represent intermediate phenotypes of disease, and variability in protein sequence or abundance is directly linked to disease mechanisms and phenotypes. Genetic association studies at the protein level provide functional evidence to map disease associations and translate this knowledge into clinical applications.
Therefore, we expect to examine the relationship between disease-causing genes and phenotypic variation at the protein level to reveal disease-related pathways, new drug targets, and translational biomarkers.
With the upgrading of mass spectrometers, proteomics technologies, and the continuous development of analysis software and databases, genome-wide and population-scale analysis of proteomic data can be achieved.
Our Discovery Proteomics Service
Our Discovery Proteomics platform enables the identification and quantification of thousands of proteins and can screen out genetic association proteins.
The discovery proteomics platform uses DIA MS proteomics methods such as swath-MS, a massively parallel and highly reproducible protein quantification technique. This combining proteomics and genome-wide association studies (PWAS) technique can be used to identify proteins that bind specifically to alleles and assess the association between abundance distribution and phenotype of each quantified protein. Thus, support unraveling the molecular mechanisms by which SNPs may play a role in gene regulation.
Benefit fields
Genetic disease research
Drug research for hereditary diseases
Workflow
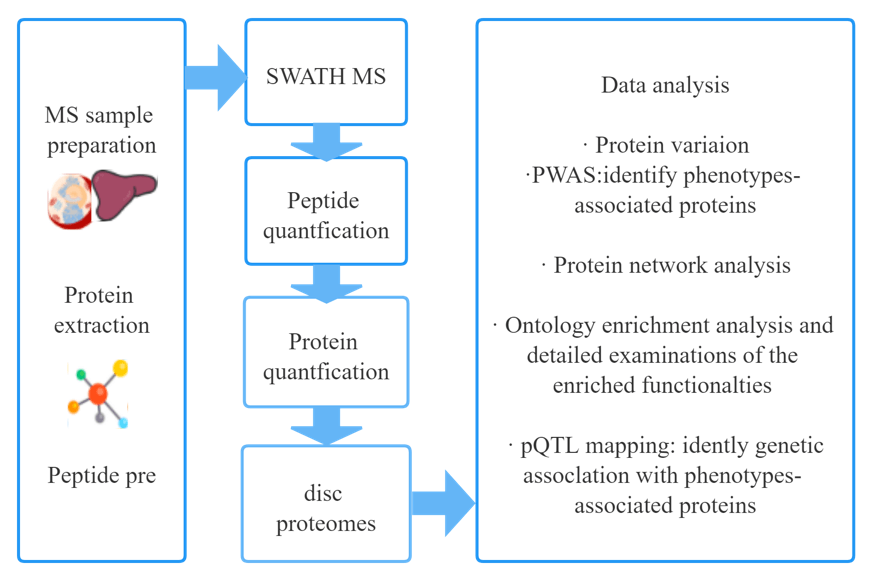
Advantages
High accuracy
High throughput, more than 9000 proteins can be identified at once
Quantitatively identify nearly all detectable molecules, covering low-abundance proteins/peptides
High repetition rate
Complete and comprehensive sample information storage in the first analysis
Sample Requirements
Animal and clinical tissue specimens: 200 mg/sample
Serum, plasma: 200 μL/sample
Cells, microorganisms: 1×107 cells/sample
Plant tender leaves and buds: 500 mg/sample
Plant seeds, fruits: 100 mg/sample
Report
- Experimental steps
- Relevant parameters
- Mass spectrometry spectra
- Raw data
- Proteomics analysis results
References:
- Karsten Suhre et al. Genetics meets proteomics: perspectives for large population-based studies.Nature Reviews Genetics volume 22, pages19–37 (2021).
- Hirokazu Okada et al.Proteome-wide association studies identify biochemical modules associated with a wing-size phenotype in Drosophila melanogaster. Nat Commun. 2016; 7: 12649.
- Butter F, Davison L, Viturawong T, Scheibe M, Vermeulen M, Todd JA, et al. (2012) Proteome-Wide Analysis of Disease-Associated SNPs That Show Allele-Specific Transcription Factor Binding. PLoS Genet 8(9): e1002982.


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)