GPF-DIA Quantitative Proteomics Services
Data independent acquisition (DIA) is a powerful technique that can significantly improve the throughput and reproducibility of proteomics assays, and has been widely used in various biological sample assays. The classical DIA process usually relies on a DDA library generation, which requires the classification of samples into multiple fractions to be assayed separately with long cycle time and high cost. In recent years, more and more analytical methods that do not rely on DDA library generation have been created, such as DIAUmpire, PECAN, encyclopeDIA, etc.
Gas-phase fractionation (GPF) improves detection rates with DIA by injecting the same sample multiple times with tiled precursor isolation windows, allowing each injection to have narrower windows (and thus fewer co-fragmented peptides) with the same instrument duty cycle. Compared with the classical DDA library generation, it can detect more target proteins (peptides) with faster project cycle time, better quantitative results, less samples and lower labor cost.
Our GPF-DIA Quantitative Proteomics Service
Our GPF-DIA quantitative proteomics service uses gas-phase ractionation technology with high resolution, accurate mass number stability, high sensitivity and high-speed scanning. All you need to do is send us your samples and we will take care of the following, including protein extraction, proteolysis, peptide separation, mass spectrometry analysis, raw mass spectrometry data analysis, and bioinformatics analysis.
Technical advantages:
Save sample size and cycle time
More accurate protein quantification
The technical core of GPF-DIA library generation:
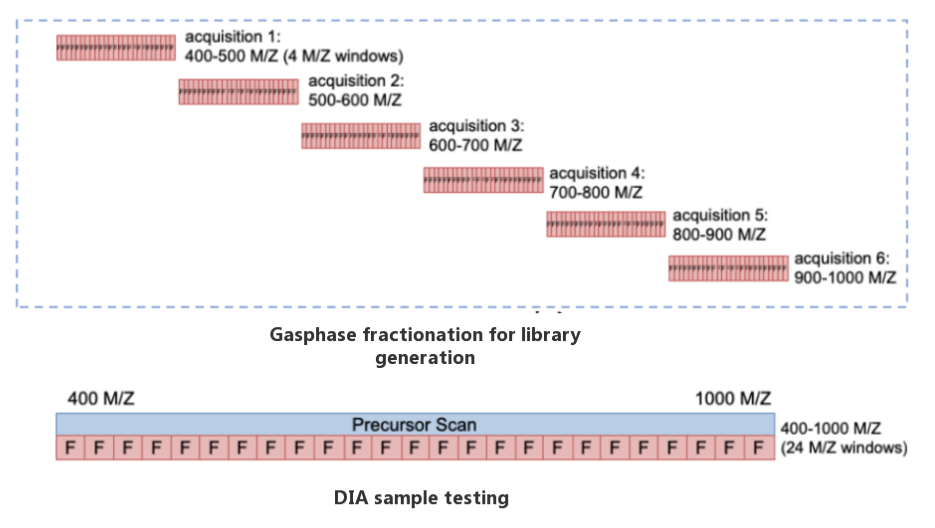
Features of GPF-DIA Library Generation:
- Scanning range of precursor ion: 400-1000m/z.
- Greatly reduce the sample size required for library generation and the cost of mass spectrometry.
- Obtain high-quality data close to the PRM.
GPF-DIA Workflow:
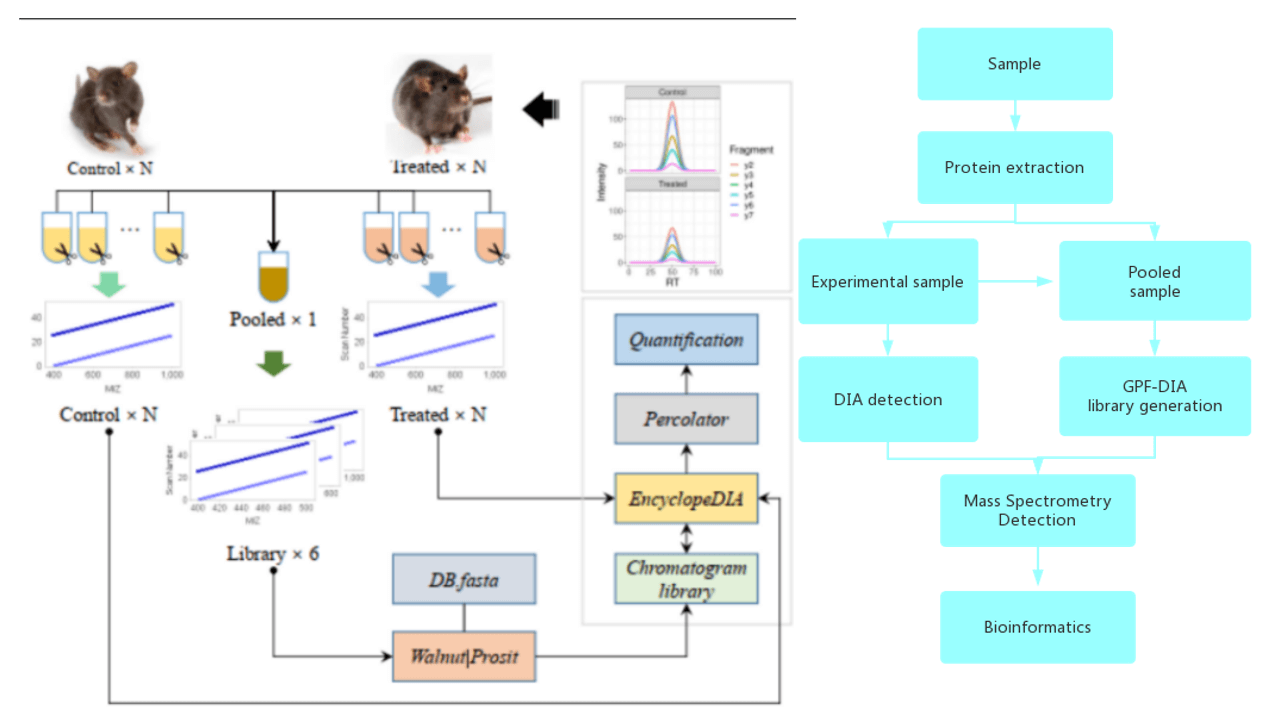
Data Analysis
| Standard data analysis content | |
| Statistical analysis of identification results | Sample protein and peptide identification histogram, PCA distribution map, quantitative variance statistical analysis |
| Bioinformatics analysis | Differential up- and down-regulated protein KEGG map, KEGG pathway function attribution, etc. |
| GO and G0 enrichment, KEGG and KEGG enrichment analysis, PPI interaction network and module analysis | |
| Advanced data analysis content | |
| Biomarker screening | Integrated machine learning, LASSO regression analysis, marker panels, ROC analysis |
| Molecular typing analysis | Unsupervised cluster typing analysis, proteome + transcriptome analysis, kinase analysis |
| Joint analysis of clinical characterization and omics results | |
| Disease-related functional module analysis | WGCNA co-expression analysis |
| Survival curve analysis | Clinical typing and survival curve analysis |
Sample Requirements:
- Animal and clinical tissue specimens: 200mg/sample
- Serum, plasma: 200μL/sample
- Cells, microorganisms: 1×107cells/sample
- Plant tender leaves and buds: 500mg/sample
- Plant seeds, fruits: 100mg/sample
Detection Platforms:

Applications:
Clinical and Pharmacology Applications:
- Disease biomarkers.
- Molecular mechanism of disease development.
- Chemical or biological drug targets.
- Novel viral inhibitors.
- Signal transduction of chemical/biological drug mechanism of action.
Agriculture and Forestry Applications:
- Improvement of meat quality and breeding conditions.
- Development mechanisms of agricultural crops.
- Research to improve food safety, taste and nutrition.
Report:
- Experimental steps
- Relevant experiment parameters
- Mass spectrometry spectra
- Raw data
- Proteomics analysis results


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)