4D Label-Free Proteomics Services
Our 4D Label-Free Proteomics Services
Using timsTOF Pro, we have launched 4D label-free proteomics services to help research in microsample proteomics and larger cohorts of clinical samples. All these services are accompanied by a high-quality full after-sales service.
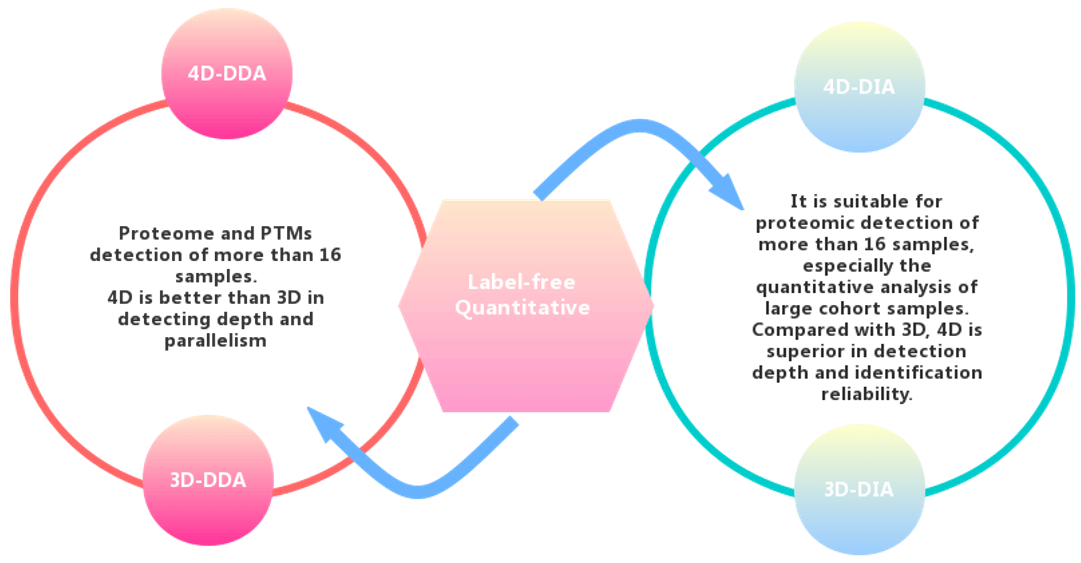
Advantages of 4D Label-Free Proteomics:
4D has greatly improved the identification accuracy, identification depth, quantification accuracy, and detection cycle, and the sample size requirements have been greatly reduced. The specific performance parameters are as follows:
1. More accurate identification
With 4 dimensional separation, the accuracy of protein identification is higher. In particular, the protein composition of biological samples is complex, and there are a large number of co-eluted peptides with small mass (m/z) differences within a small retention time. Ion mobility is used to distinguish peptides with small differences in mass (different ion mobility) to achieve more accurate identification.
2. More proteins identified
Under the same test conditions, 4D label-free has more than 50% higher detection capacity than traditional label-free technology. For example, 5200 hela proteins were identified using 4D label-free proteome in 90 min, and identified proteins increased by 62.5%. In another study, more than 500 blood proteins were identified by 4D label-free proteome in 11.5 min, and identified proteins increased by 50%.
3. More stable quantification
Continuous injection of 96 needles (R2>0.95, CV<5%), the quantitative range exceeds 3 orders of magnitude.
4. Shorter project cycle
Unique acquisition method to achieve ultra-high scanning speed and significantly shorten the experimental cycle time.
5. Lower sample requirement
The sample load is only 200ng, which can achieve deep coverage.
Applications:
- Protein expression profile of drug response, gene knockout/overexpression
- Study on the growth and development of plants and microorganisms; Study on the physiology of stress and adversity of plants and microorganisms
- High-throughput analysis of disease biomarkers and drug targets
- Protein mechanism protein and special function protein screening
- The best way to detect clinical micro-samples, such as: exosomes, FFPE paraffin sections, biopsy puncture tissues, etc.
Workflow
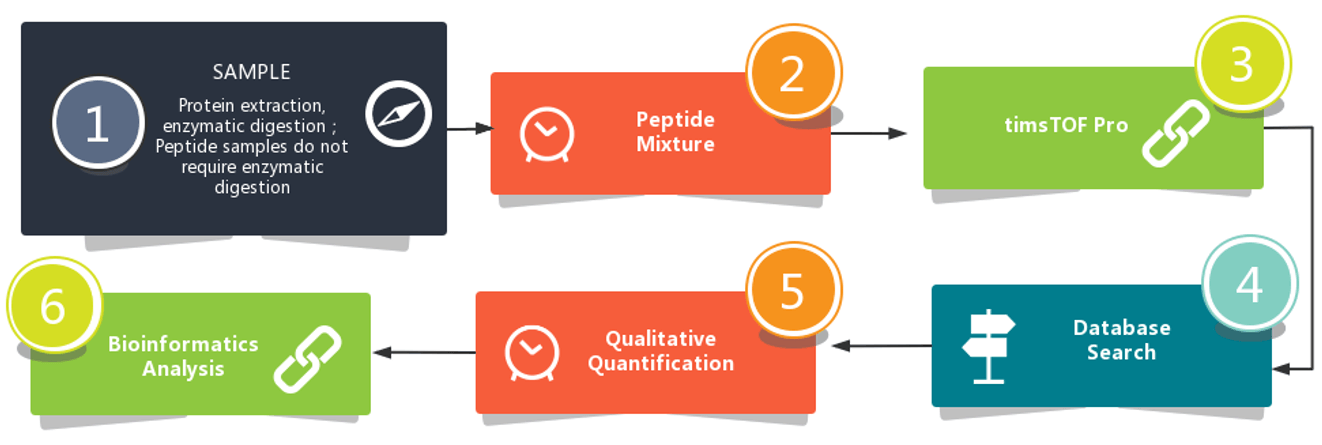
Data Analysis
| Standard data analysis content | |
| Total protein identification analysis | Protein identification and quantification, differential outcome statistics, volcano map, cluster heatmap |
| Differential protein function analysis | Subcellular localization, structural domain analysis, GO function analysis, KEGG pathway analysis, PPT interaction network analysis |
| QC | Peptide ion mass deviation distribution, peptide ion score distribution, protein abundance ratio distribution |
| Advanced data analysis content | |
| WGCNA co-expression analysis | Obtain core proteins strongly associated with phenotype |
| Cluster trend analysis | Explore protein expression trend |
Sample Requirements:
- Co-IP Samples: Protein requirement is around 2-5ug, do not include decontaminants (e.g. SDS) and high oxygen concentration in the sent protein samples; or you can send the beads containing protein samples to us directly and we will take care of the subsequent experiments.
- Tissue Samples: Please send us the tissue samples on dry ice.
- Protein Samples: Please make sure the total amount of protein is above 50ug; ordinary tissue and cell lysate can be used for protein extraction.
- Transportation: Please use sufficient amount of dry ice for transportation and try to use a fast postal delivery method to reduce the possibility of sample degradation during transportation.
| Tissue Samples | Protein | # of Cells | Animal Tissue | Plant Tissue | Blood | Urine | Serum | Microbes |
| Quantify | 100 ug | 1×107 cells | >200 mg | 1 g | 1 mL | 2 mL | 0.2-0.5 mL | Dry weighed: 200 mg |
Report:
- Experimental steps
- Relevant experimental parameters
- Mass spectrometry spectra
- Raw data
- Proteomics analysis results
In addition, we also provide 4D DIA Proteomics and 4D PTMs Proteomics services. Please contact us for more details.


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)