4D-DIA Quantitative Proteomics Services
4D-DIA proteomics is a next-generation DIA technology based on the timsTOFPro mass spectrometer for differential quantitative proteomics analysis. 4D-DIA proteomics scanning is faster and more sensitive compared to 3D-DIA technology. Meanwhile, the addition of the fourth-dimensional ion drip separation allows DIA data to be acquired without sacrificing window cycling speed while reducing spectral complexity and improving ion utilization, achieving a comprehensive improvement in proteomics in terms of coverage depth, sensitivity, and throughput.
Principle of DIA
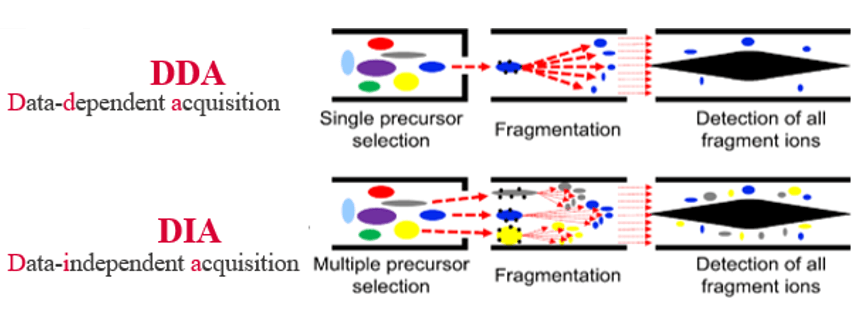
Proteomics techniques can be divided into data-dependent acquisition (DDA) strategies and data-independent acquisition (DIA) strategies.
- DDA strategy: a limited number of peptide ions with higher intensities are acquired in each time window in the primary mass spectrometry and enter the secondary mass spectrum for fragmentation analysis.
- DIA strategy: the whole full scan range of the mass spectrum is divided into several windows and all ions in each window are selected, fragmented and detected cyclically.
- The DIA technique can obtain fragmentation information of all ions in the sample. Therefore, more quantitative information can be obtained, and the quantification is more accurate and reproducible based on the MS2 spectra.
Our 4D-DIA Proteomics Services:
Using timsTOF Pro, we have launched 4D-DIA proteomics services to help research in microsample proteomics, large cohort of clinical samples, and high-throughput protein analysis.
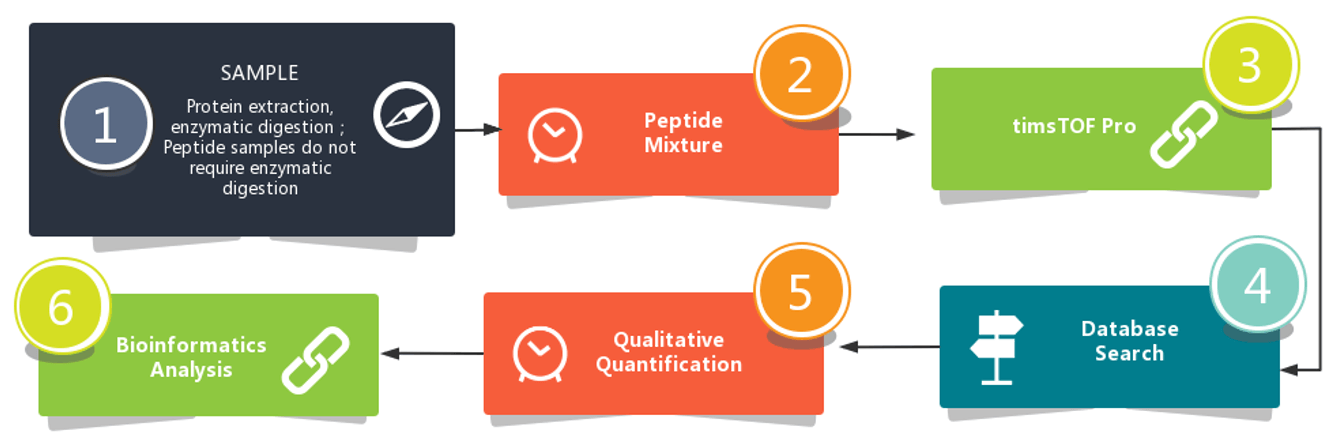
Workflow
Data Analysis
| Standard data analysis content | |
| Statistical analysis of identification results | Sample protein and peptide identification histogram, PCA distribution map, quantitative variance statistical analysis |
| Bioinformatics analysis | Differential up- and down-regulated protein KEGG map, KEGG pathway function attribution, etc. |
| GO and G0 enrichment, KEGG and KEGG enrichment analysis, PPI interaction network and module analysis | |
| Advanced data analysis content | |
| Biomarker screening | Integrated machine learning, LASSO regression analysis, marker panels, ROC analysis |
| Molecular typing analysis | Unsupervised cluster typing analysis, proteome + transcriptome analysis, kinase analysis |
| Joint analysis of clinical characterization and omics results | |
| Disease-related functional module analysis | WGCNA co-expression analysis |
| Survival curve analysis | Clinical typing and survival curve analysis |
Advantages:
- Nearly 100% ion utilization rate greatly improves detection sensitivity
- Significantly improve the detection depth
- Comprehensively improve quantitative integrity
Application:
- Larger cohorts of clinical sample analysis, such as precision typing of major diseases such as cancer and cardiovascular diseases.
- The best way to detect clinical microsamples, such as: exosomes, FFPE paraffin sections, biopsy punctured tissues, etc.
- Large-scale sample research on crop population traits, such as breeding lines, stress intervention, etc.
- Clinical biomarker discovery: DIA is close to the quantitative ability of MRM, realizing the integration of marker screening and preliminary verification process.
- Biological sample information database construction: DIA enables complete preservation of biological information for subsequent retrospective analysis such as drug response and protein expression profiling of gene knockout/overexpression.
Sample Requirements:
- Co-IP Samples: Protein requirement is around 2-5ug, do not include decontaminants (e.g. SDS) and high oxygen concentration in the sent protein samples; or you can send the beads containing protein samples to us directly and we will take care of the subsequent experiments.
- Tissue Samples: Please send us the tissue samples on dry ice.
- Protein Samples: Please make sure the total amount of protein is above 50ug; ordinary tissue and cell lysate can be used for protein extraction.
- Transportation: Please use sufficient amount of dry ice for transportation and try to use a fast postal delivery method to reduce the possibility of sample degradation during transportation.
| Tissue Samples | Protein | # of Cells | Animal Tissue | Plant Tissue | Blood | Urine | Serum | Microbes |
| Quantify | 100 ug | 1×107 cells | >200 mg | 1 g | 1 mL | 2 mL | 0.2-0.5 mL | Dry weighed: 200 mg |
Report:
- Experimental steps
- Relevant experimental parameters
- Mass spectrometry spectra
- Raw data
- Proteomics analysis results
In addition, we also provide 4D Label-Free Proteomics and 4D-PTMs Proteomics services. Please contact us for more details.


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)