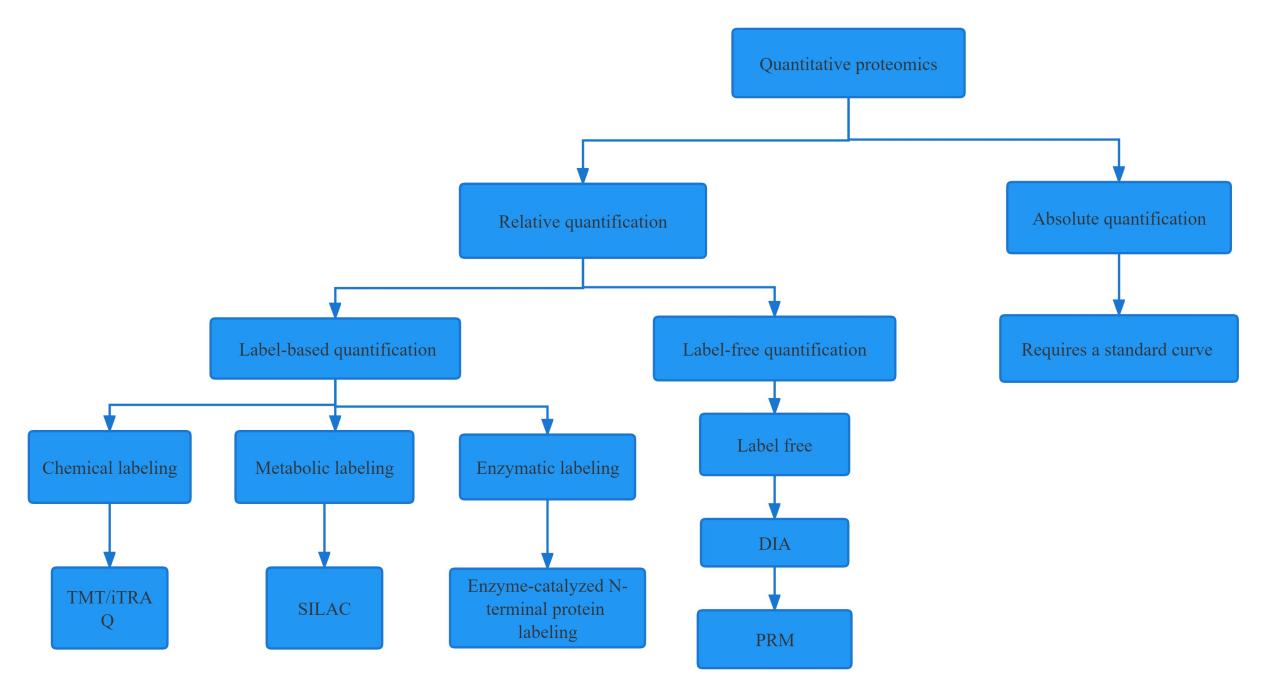
Differences Between DIA, TMT/iTRAQ, And Traditional Label-free
In recent years, biological mass spectrometry technology has been developed rapidly, especially in mass spectrometry. With this progress, a large number of experimental techniques that can be combined with biological mass spectrometry have emerged, such as DIA (Data independent acquisition), TMT/iTRAQ, and label-free.
This article explores the unique strengths and capabilities of these three popular technologies to help researchers choose the most appropriate technical route. DIA is an emerging technique admired for its ability to analyze large numbers of peptides simultaneously, making it ideal for quantitative analysis of complex proteomes. On the other hand, TMT/iTRAQ and label-free technologies have existed for some time and are considered to be old-fashioned and powerful. TMT/iTRAQ can perform multiplex quantification of up to 10 or more samples simultaneously, making it popular for high-throughput research. On the other hand, label-free is a simpler and more economical method that relies on spectral counts to quantify protein abundance.
Summary of Proteomics Techniques
| Technology Type | DIA | TMT | Traditional Label free |
| Sample Throughput | Large, especially suitable for large-scale clinical samples | Smaller, suitable for projects with fewer samples (highest throughput of TMT currently reaches 16 labels) | Large, single needle injection |
| Quantitative Stability | High, fewer missing values | High, fewer missing values | Low, more missing values |
| Identification Coverage | High, with full-scan coverage in windows | Relatively high, with separation of multiple components after labeling | Low |
| Reproducibility | High | High | Low |
| Cost | Medium | High | Low |
| Detection Sensitivity | High | High | Low |
Compare of DIA, TMT/iTRAQ, and Label free
Label free
Label free refers to the direct identification and qualitative and quantitative analysis of peptide segments without using any labeling reagents. The experimental process is simple, and only requires conventional enzymatic digestion and desalination of the protein prior to mass spectrometry analysis. There are two methods for quantitative analysis: peak intensity and spectral count, with peak area being the most commonly used. However, due to the fact that only the top N precursor ions are selected for secondary fragmentation and MS2 detection during mass spectrometry acquisition, some information on low-abundance peptide segments will be lost, resulting in a high number of missing values.
TMT/iTRAQ Labeled Quantitative Proteomics
TMT is short for Tandem Mass Tag, which is a classic chemical labeling reagent developed by Thermo Fisher Scientific. It is widely used in the analysis of differentially expressed proteins in proteomics research. This technology uses 6-plex, 10-plex, or 16-plex isotopic labels, which covalently bind to the amino groups of peptide segments and enable qualitative and quantitative analysis of proteins in 6/10/16 different samples simultaneously.
The TMT reagent consists of three chemical groups: a reporter group, a balance group, and a reactive group. The reactive group specifically binds covalently to the amino group of peptide segments, linking the TMT reagent to the peptide.
In the MS1 spectrum, the same peptide of different samples, labeled with the same complete TMT reagent, appear as a single peak. However, inside the collision cell, the balance group undergoes neutral loss, while the reporter group generates corresponding reporter ions. The intensity of these reporter ions represents the intensity of the corresponding sample protein/peptide, enabling simultaneous quantification of 6/10/16 different samples.
The TMT labeling technology is stable and has good reproducibility, but the commercialized reagent kits are expensive and the number of labeling channels is limited, making it unsuitable for studying a large number of samples.
DIA Proteomics
Data independent acquisition (DIA) is one of the highly regarded mass spectrometry acquisition technologies in recent years, which is also a type of label-free technology. Compared to traditional label-free, the greatest advantage of DIA is its ability to efficiently measure low-abundance protein molecules in complex samples, greatly improving the reliability of quantitative analysis.
The principle of DIA technology is to perform full scanning on all precursor ions and their fragment ions in each acquisition window at a high speed and cyclically during mass spectrometry data acquisition, rather than selecting them based on precursor ion signal intensity and then performing secondary fragmentation and scanning.
DIA has the advantages of high throughput, high resolution, high reproducibility, and accurate quantification. Moreover, samples do not need to be graded, greatly shortening the detection time for each sample, making it suitable for large-scale proteomic studies.
References:
- Zhu, W., J. Smith and C. Huang. "Mass Spectrometry-Based Label-Free Quantitative Proteomics." Journal of Biomedicine and Biotechnology 2010 (2010): n. pag.
- Gupta, Meera & Sonnett, Matthew & Ryazanova, Lillia & Presler, Marc & Wühr, Martin. (2018). Quantitative Proteomics of Xenopus Embryos I, Sample Preparation: Methods and Protocols. 10.1007/978-1-4939-8784-9_13.
- Pappireddi, Nishant & Martin, Lance & Wühr, Martin. (2019). A Review on Quantitative Multiplexed Proteomics. ChemBioChem. 20. 10.1002/cbic.201800650.


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)