Isolation, Characterization, Single-Particle Phenotyping (ExoView/SP-IRIS) & Multi-Omics
Creative Proteomics provides comprehensive exosome analysis services for biomarker research, engineered exosome development, and research-stage process trending. We specialize in delivering end-to-end exosome characterization, phenotyping, and downstream analysis solutions, supporting both academic research and R&D teams. Our services are designed to meet your specific needs, from sample isolation and purification to advanced single-particle phenotyping and multi-omics analysis.
Exosomes are a subtype of extracellular vesicles (EVs). If you require broader EV workflows, please refer to our Extracellular Vesicles (EV) Analysis Services.
Applications Capabilities Method Selection Workflow Sample Types Deliverables FAQs Get a Custom Proposal
Our Exosome Analysis Services Solve Key Research Challenges
We provide high-quality, reproducible results that support diverse research goals, whether you're investigating biofluid biomarkers, developing engineered exosomes, or monitoring research-stage process consistency. Our services are flexible and tailored to each project's specific requirements.
Biomarker Discovery in Biofluids
Challenge: Isolating and identifying exosome biomarkers from complex biofluids like plasma, serum, and CSF can be challenging due to background interference and low abundance.
Solution:
- Exosome isolation & purification using proven methods like polymer-based enrichment, SEC, and immunomagnetic beads.
- Marker verification with WB, TEM, and NTA to confirm exosome presence and quality.
- Subpopulation profiling with ExoView for accurate quantification of marker-defined subsets.
- Reliable longitudinal tracking for cohort studies.
Engineered Exosome Development & Therapeutics
Challenge: Optimizing engineered exosomes for targeted delivery and ensuring efficient cargo loading and surface phenotyping.
Solution:
- Exosome engineering: Surface ligand optimization and multi-marker profiling.
- Cargo loading validation and uptake assays to verify therapeutic delivery.
- Single-particle phenotyping (ExoView) for cargo-positive quantification and subpopulation analysis.
- Functional readouts for therapeutic efficacy evaluation.
Process Development & Research QC
Challenge: Maintaining batch-to-batch consistency and ensuring process quality in exosome production.
Solution:
- Standardized panels for reliable process monitoring.
- ExoView profiling to track exosome subset proportions and ensure reproducibility.
- QC reporting to ensure sample consistency and minimize bias across experiments.
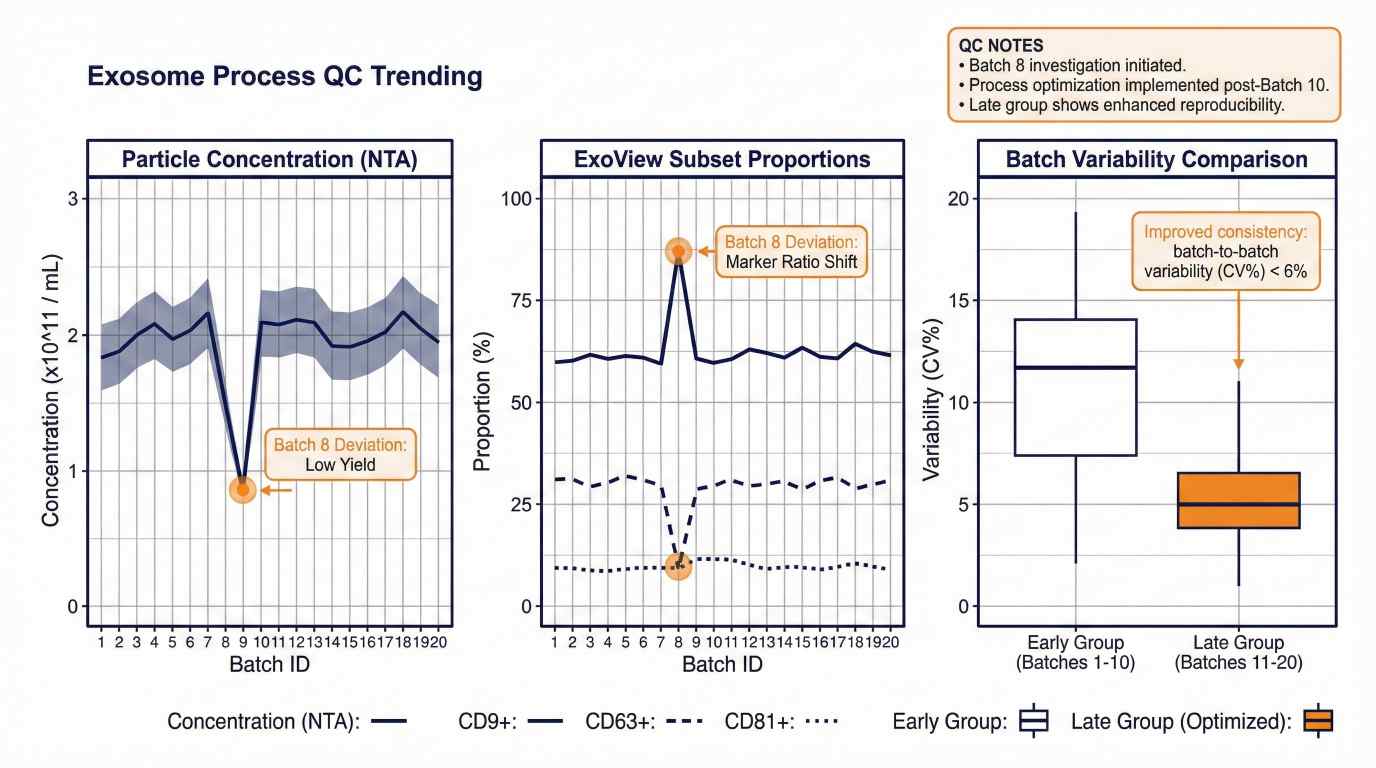 Biofluid exosome biomarker discovery workflow: isolation, characterization (NTA/TEM/WB), and ExoView subpopulation profiling.
Biofluid exosome biomarker discovery workflow: isolation, characterization (NTA/TEM/WB), and ExoView subpopulation profiling.
Exosome Analysis Capabilities
We offer several isolation techniques to meet your specific needs, from high-throughput sample processing to optimized isolation for downstream applications.
- Sucrose Gradient / Density Centrifugation
- Polymer-Based Enrichment
- Immunomagnetic Bead Enrichment
- Size Exclusion Chromatography (SEC)
Typical outputs: Clean, isolated exosome fractions for downstream applications such as characterization, phenotyping, and multi-omics analysis.
Exosome Identification and Characterization
For baseline validation and characterization of exosome samples, we provide the following services:
Nanoparticle Tracking Analysis (NTA)
- Best for: Baseline concentration and size distribution trending.
- Typical outputs: Particle concentration, size distribution histogram.
Transmission Electron Microscopy (TEM)
- Best for: Visual confirmation and morphology analysis of isolated exosomes.
- Typical outputs: High-resolution TEM images.
Exosome Marker Verification (Western Blot / Immunoassay)
- Best for: Confirming the presence of canonical exosome markers (e.g., CD9, CD63, CD81).
- Typical outputs: Presence/absence of markers with comparative signal documentation.
Exosome Marker Analysis (Panel-Based)
- Best for: Targeted marker validation and characterization of exosome populations.
- Typical outputs: Marker-focused results with interpretation notes (project-dependent).
Single-Particle Exosome Phenotyping (ExoView / SP-IRIS)
For studies that require a deeper understanding of exosome subpopulations, co-expression, and cargo association, we offer ExoView single-particle profiling.
- Best for: Identifying and quantifying exosome subpopulations at the single-vesicle level.
- Services:
- Marker-defined subpopulation proportions (e.g., CD9/CD63/CD81)
- Multi-marker co-expression patterns via fluorescence colocalization (design-dependent)
- Exportable, publication-ready figures and data tables for analysis
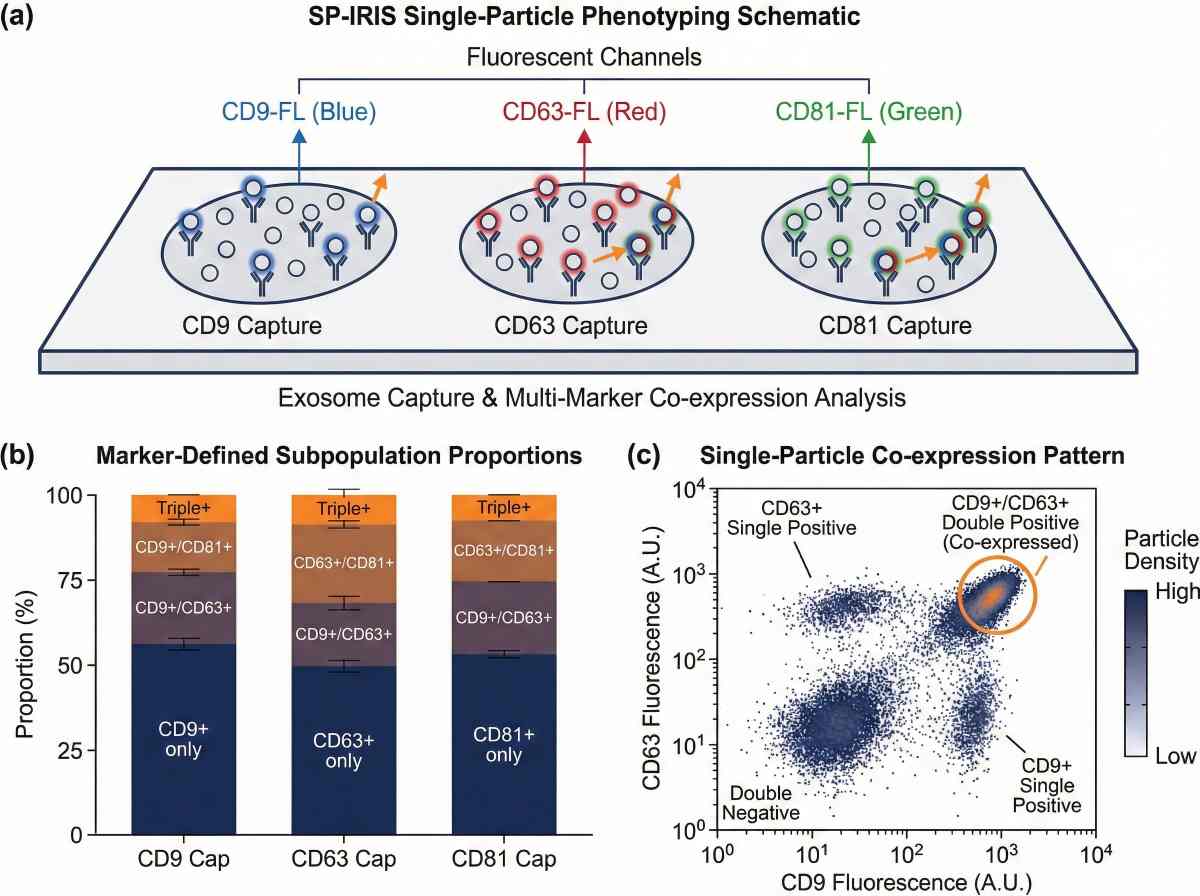 ExoView / SP-IRIS single-particle exosome phenotyping outputs for subpopulation quantification and co-expression analysis.
ExoView / SP-IRIS single-particle exosome phenotyping outputs for subpopulation quantification and co-expression analysis.
Exosome Engineering and Therapeutics Support
For engineered exosome development and therapeutic research, we provide services to support targeted delivery systems, surface phenotype verification, and cargo loading.
Optional Multi-Omics Add-Ons (Mechanistic Discovery)
When deeper pathway-level insight is needed, we offer optional omics modules (availability depends on sample type and study goals).
- Exosome Proteomics (discovery or targeted, project-dependent)
- Exosome Metabolomics (project-dependent)
Best for: Mechanistic discovery, pathway mapping, and functional insight generation.
How to Choose the Right Exosome Analysis Method
| Research Goal |
Recommended Method |
Why It's Used |
| Baseline screening (size/count) |
NTA |
Quick concentration and size distribution |
| Morphology confirmation |
TEM |
Visual documentation of exosome structure |
| Marker confirmation |
WB / Immunoassay |
Validation of canonical exosome markers |
| Subpopulations & co-expression |
ExoView (single-particle) |
Resolves heterogeneity and co-expression |
| Mechanistic discovery |
Multi-Omics (Proteomics/Metabolomics) |
Pathway and functional insights (optional) |
Typical Project Workflow for Exosome Analysis
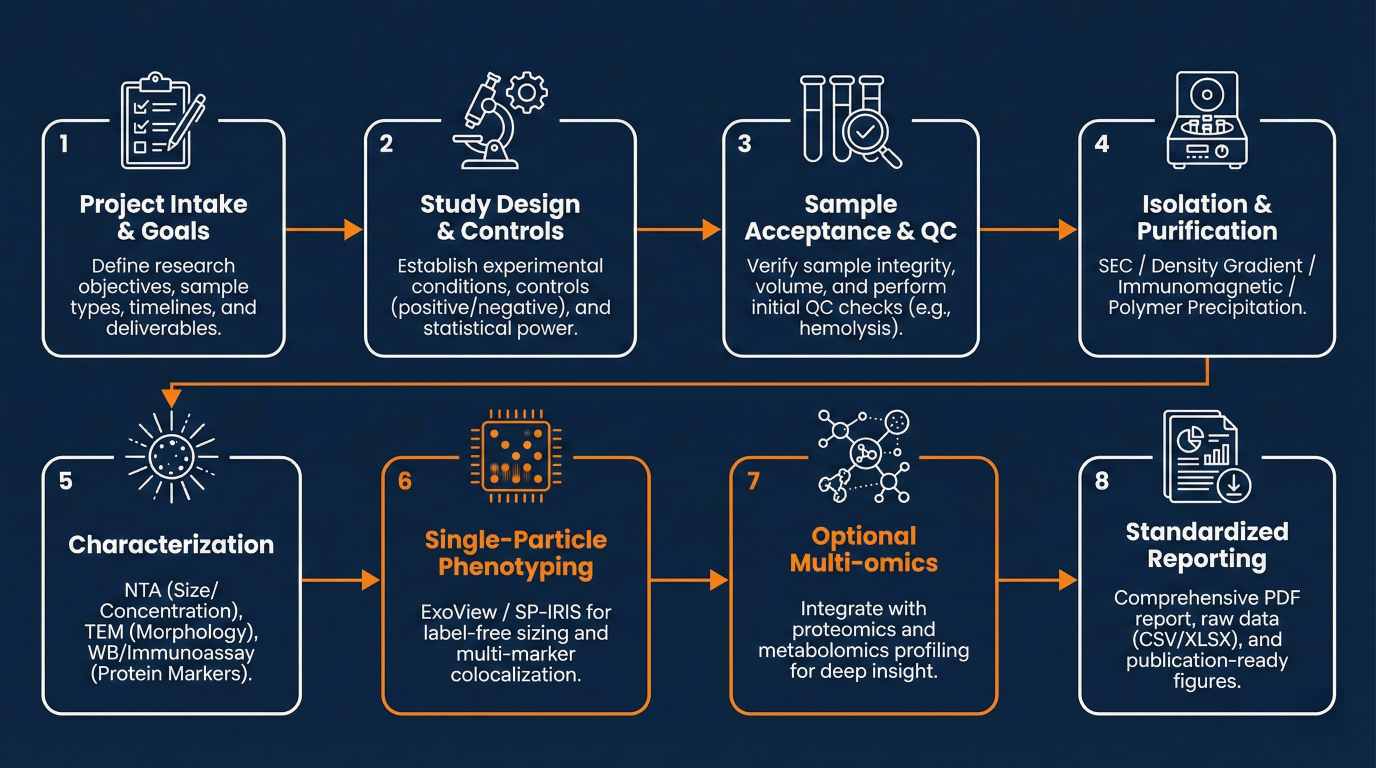 End-to-end exosome analysis workflow from isolation and characterization to ExoView phenotyping and reporting.
End-to-end exosome analysis workflow from isolation and characterization to ExoView phenotyping and reporting.
Supported Exosome Sample Types
We support a range of biofluid and cell culture samples for exosome analysis. Exact requirements will be confirmed during feasibility planning.
| Sample Type |
Typical Use Case |
Notes |
| Cell culture supernatant |
Engineering/process studies |
Media/serum use, processing history |
| Plasma / Serum |
Biomarker discovery, therapeutic R&D |
Anticoagulant, hemolysis/lipemia notes |
| Urine |
Non-invasive biomarker studies |
Collection timing and handling notes |
| CSF |
Low-abundance workflows |
Metadata and careful control design |
| Other biofluids |
Case-by-case |
Feasibility reviewed before start |
Sample Submission Checklist
To maximize success and comparability, please include:
- Sample type, volume, and matrix details (e.g., plasma/serum/CSF/urine; culture conditions)
- Collection and storage conditions (temperature, freeze–thaw cycles, processing time)
- Key metadata (cohort groups, timepoints, treatment conditions)
- Known confounders (e.g., hemolysis, lipemia, anticoagulant type)
- Study goal (biomarker discovery vs. engineering vs. QC trending) and preferred readouts
Exosome Analysis Deliverables and Reporting
Standard deliverables
- Standardized Report (PDF): Methods, QC notes, interpretation guidance
- Data Tables (CSV/XLSX): For downstream statistics and cohort comparisons
- Publication-Ready Figures: High-resolution exports for publications and presentations
Optional deliverables
- Figure Pack: Ready-to-use figures for manuscripts/grants
- Study Design Memo: Including panel/controls/replicate plan
- Add-On Omics Data: When multi-omics services are selected
FAQs – You May Want to Know
What is the best method to isolate exosomes (SEC vs polymer precipitation vs immunoaffinity)?
There isn't one "best" method—choose based on your priority. SEC is often preferred when downstream functional assays or omics need higher purity and reproducibility. Polymer precipitation can maximize yield but may co-enrich proteins/lipoproteins that interfere with assays. Immunoaffinity/immunomagnetic capture increases specificity for marker-defined vesicles but can bias toward captured subpopulations and reduce total yield. A practical rule: purity-sensitive readouts → SEC; rapid enrichment/yield → polymer; marker-specific questions → immunoaffinity.
How do you confirm that what I have are exosomes and not contaminants?
Use converging evidence: particle metrics, morphology, and marker data plus at least one negative marker. A common approach is multiple positive EV proteins (CD9/CD63/CD81 plus intraluminal proteins such as TSG101/ALIX/HSP70) and a negative marker (often calnexin, sample-dependent) to flag cellular contamination. Combining these reduces false positives from protein aggregates or debris.
Which exosome markers should I test for Western blot?
A robust WB set typically includes 2–3 surface markers (commonly CD9, CD63, CD81), 1–2 intraluminal markers (e.g., TSG101, ALIX, HSP70), and at least one negative control (often calnexin, depending on sample). Using more than one marker class improves interpretability and cross-study comparability.
Can ExoView / SP-IRIS replace NTA for counting exosomes?
They answer different questions. NTA estimates particle concentration and size distribution for the total particle population but cannot phenotype individual vesicles. ExoView/SP-IRIS is antibody-capture single-particle phenotyping: it quantifies marker-defined captured subsets and co-expression patterns, so results depend on panel design and capture biology. Many studies use NTA for global trending and ExoView for phenotype-specific quantification.
What does SP-IRIS actually measure in ExoView assays?
SP-IRIS detects and counts individual nanoparticles captured on an antibody array using interferometric imaging, and can overlay fluorescence to assess marker co-expression. It is strongest for counts and phenotypes within captured marker subsets, not for an unbiased census of all EVs in the sample.
Why do different isolation methods change my biomarker or omics results?
Isolation changes the purity–yield balance and co-isolated background (especially in plasma/serum where abundant proteins and lipoproteins are present). These differences can shift apparent biomarker signals and affect proteomics/metabolomics sensitivity and missingness. Standardizing one workflow, documenting metadata, and keeping controls consistent are key to interpretable cohort comparisons.
Can you work with low-volume or low-abundance samples like CSF?
Yes, but low-abundance matrices require careful feasibility planning and stronger controls (process blanks, consistent handling metadata, and replicates where possible). Workflows should prioritize background reduction and sensitive readouts aligned to the claim you need (e.g., phenotype shift vs mechanistic discovery).
What controls should I include for a cohort biomarker study?
Yes, but low-abundance matrices require careful feasibility planning and stronger controls (process blanks, consistent handling metadata, and replicates where possible). Workflows should prioritize background reduction and sensitive readouts aligned to the claim you need (e.g., phenotype shift vs mechanistic discovery).
How do I choose between total exosome profiling and subpopulation profiling?
Use total readouts when you need global trending across many samples or conditions. Use subpopulation profiling when biology depends on cell-of-origin markers, co-expression, or cargo association (common in engineered exosomes and disease heterogeneity). Single-particle antibody-capture methods are designed to resolve this heterogeneity.
Do I need to use the same isolation method across all samples in a study?
Usually yes for cohort comparisons and process trending. Switching isolation methods can introduce systematic shifts in purity, yield, and background carryover that mimic biological differences. If a change is unavoidable, run a bridging set (the same samples processed by both methods) to quantify the shift and preserve interpretability.



 Biofluid exosome biomarker discovery workflow: isolation, characterization (NTA/TEM/WB), and ExoView subpopulation profiling.
Biofluid exosome biomarker discovery workflow: isolation, characterization (NTA/TEM/WB), and ExoView subpopulation profiling. ExoView / SP-IRIS single-particle exosome phenotyping outputs for subpopulation quantification and co-expression analysis.
ExoView / SP-IRIS single-particle exosome phenotyping outputs for subpopulation quantification and co-expression analysis. End-to-end exosome analysis workflow from isolation and characterization to ExoView phenotyping and reporting.
End-to-end exosome analysis workflow from isolation and characterization to ExoView phenotyping and reporting.