Cell surface proteomics helps you measure proteins exposed on the plasma membrane. This surfaceome profiling service is designed for teams who need evidence of target accessibility, not just gene expression. You get LC–MS/MS data that supports drug target discovery, biomarker screening, and cell phenotype comparison.
Surface proteins often change with cell state, disease context, or treatment. Direct measurement helps reduce false positives that can appear when you infer localisation from transcriptomics or bulk proteomics alone.
Service Highlights Applications Platform Workflow Experimental Design Sample Requirements Deliverables CaseFAQ
Cell Surface Proteomics: What It Measures
Cell surface proteomics measures proteins presented on the plasma membrane and exposed to the extracellular space. This surfaceome profiling service helps you bridge the gap between "expression signals" and evidence of surface accessibility, which is critical for antibody-based programs and actionable biomarker discovery.
Workflows combine surface-focused enrichment with LC–MS/MS and optional DIA (data-independent acquisition) quantitative proteomics to support consistent comparisons across conditions.
Cell Surface Proteomics Service Highlights for Target Discovery and Biomarkers
- Surface-focused enrichment designed to prioritise extracellular exposure
- LC–MS/MS identification with optional DIA quantitative surfaceome profiling
- Multiple enrichment routes to match biology and sample constraints
- Membrane-aware sample preparation options for difficult hydrophobic proteins
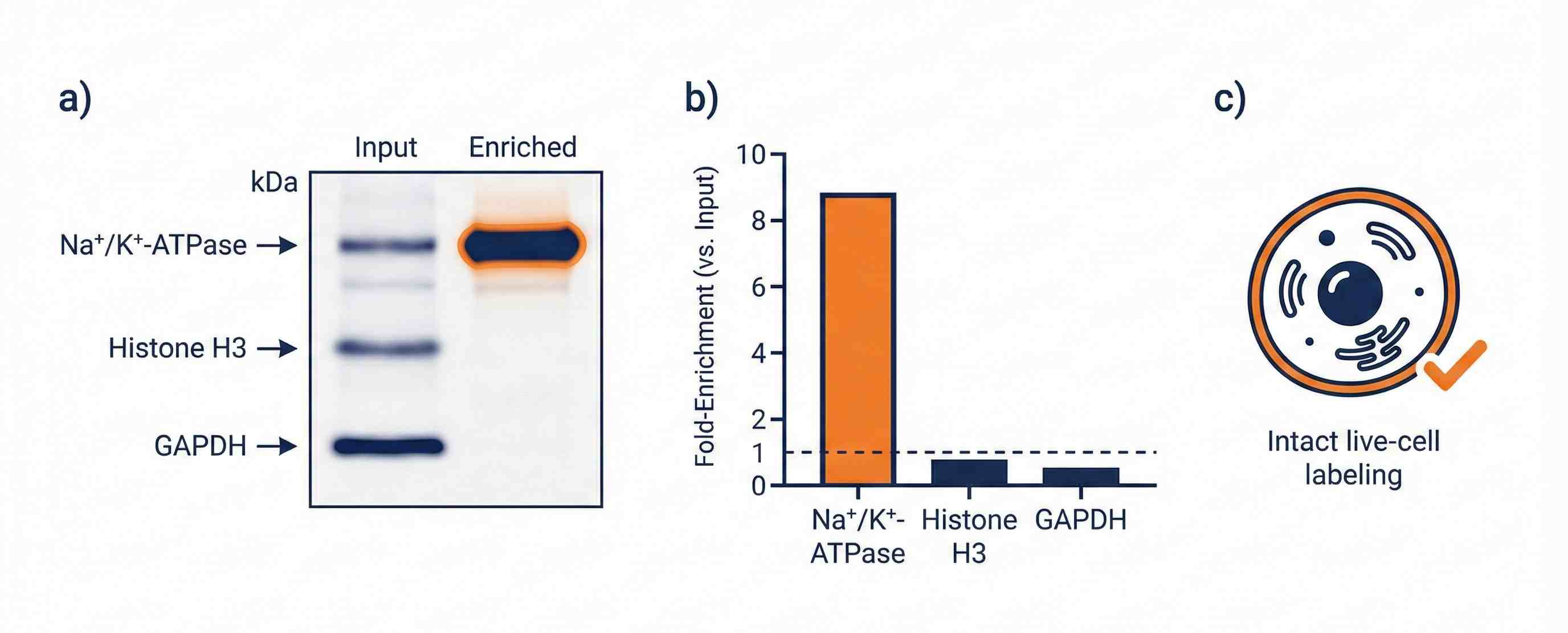
- QC summaries to assess enrichment specificity and membrane integrity
Surfaceome Profiling Applications and Use Cases (Discovery to Lead Optimisation)
Surfaceome profiling is used when surface accessibility and model relevance drive decisions. Cell surface proteins include receptors, transporters, and adhesion molecules, and their presentation can shift with disease state, differentiation, and treatment.
Goal-to-solution guide
| Goal |
What surfaceome data can support |
Common next steps |
| Therapeutic target discovery |
Evidence of extracellular exposure and condition-specific changes |
Flow cytometry, IHC/IF, functional assays |
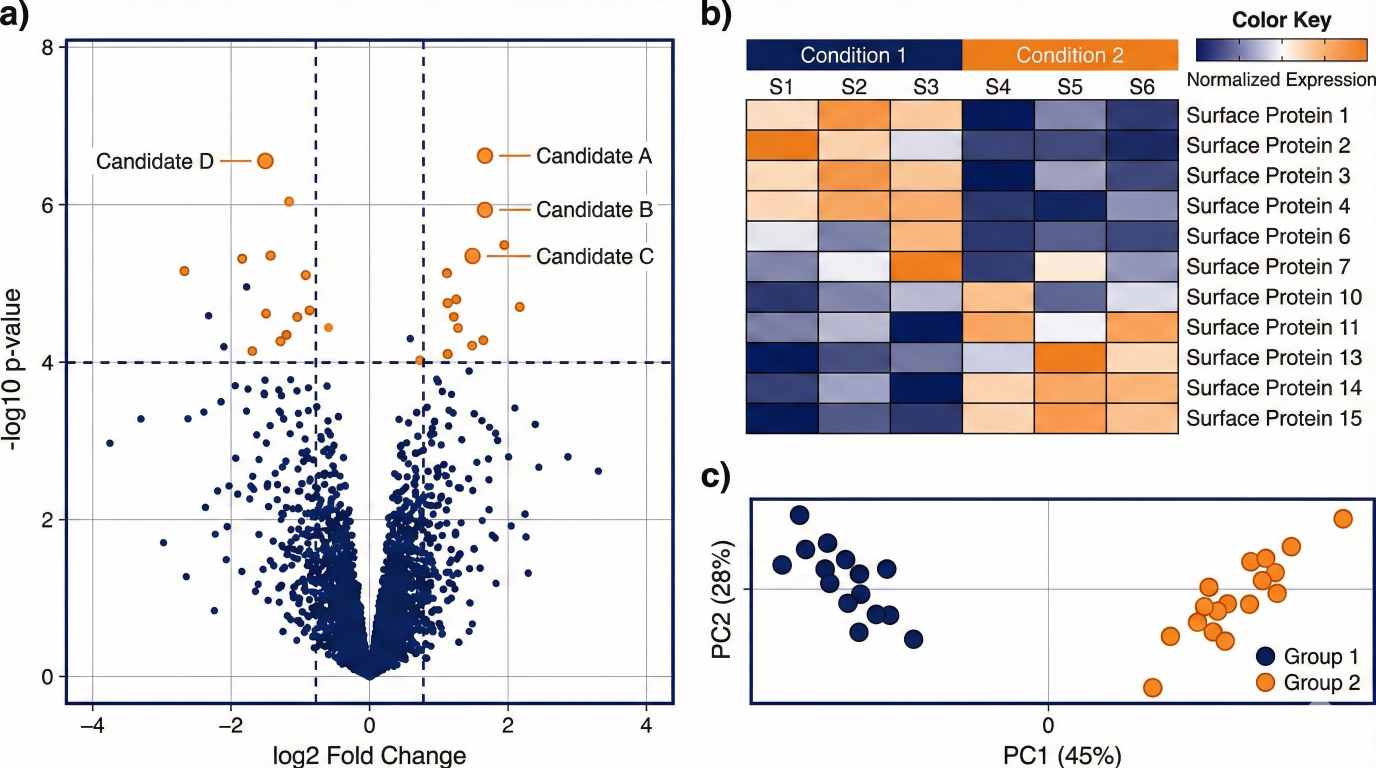
| Biomarker discovery |
Marker candidates that separate phenotypes across groups |
Panel design, assay development planning |
| Cell phenotyping and model selection |
Surface marker patterns across cell lines or primary research samples |
Cell sorting strategy, model selection |
| Drug-response and MOA studies |
Treatment-driven surfaceome remodelling signals |
Mechanism follow-up, targeted validation |
ADC and bispecific target prioritisation (extracellular domain focus)
Surfaceome data supports shortlisting candidates with evidence of extracellular exposure. Where appropriate, results can be reviewed at the peptide level to support extracellular-domain (ECD) evidence and reduce the risk of pursuing targets that are primarily internal.
Antigen screening for cell therapy (CAR-T/NK and related programs)
Surface profiling helps identify antigens measurable on the cell exterior and supports condition-to-condition comparisons. It can also help prioritise discovery-stage candidates for follow-up safety and specificity assessments.
Cell surface biomarker discovery for stratification and translational readouts
Surfaceome datasets can reveal marker candidates that separate phenotypes and are compatible with antibody-based assays. This can support development of practical validation panels and translational readouts.
Treatment-induced surfaceome changes (research only)
Drug exposure can remodel surface presentation. Surfaceome profiling can support comparisons such as treated vs baseline to help identify resistance markers, compensatory pathways, or combination hypotheses in cell models.
Advantages of Our Cell Surface Proteomics Service
Surface-Accessibility Evidence
Enrichment is designed to prioritise extracellular exposure, not total cellular abundance.
Fit-for-Purpose Enrichment Options
Biotinylation, trypsin shaving, or glyco-capture can be matched to your biology and constraints.
Membrane-Protein Aware Prep
Preparation strategies are built for hydrophobic and multi-pass targets to support LC–MS/MS recovery.
DIA-Ready Quantification
Optional DIA supports consistent comparisons across conditions when quantification is central.
QC for Specificity and Integrity
Marker-based checks help assess enrichment specificity and reduce intracellular carryover risk.
Decision-Ready Deliverables
Outputs are structured for technical review, prioritisation, and downstream validation planning.
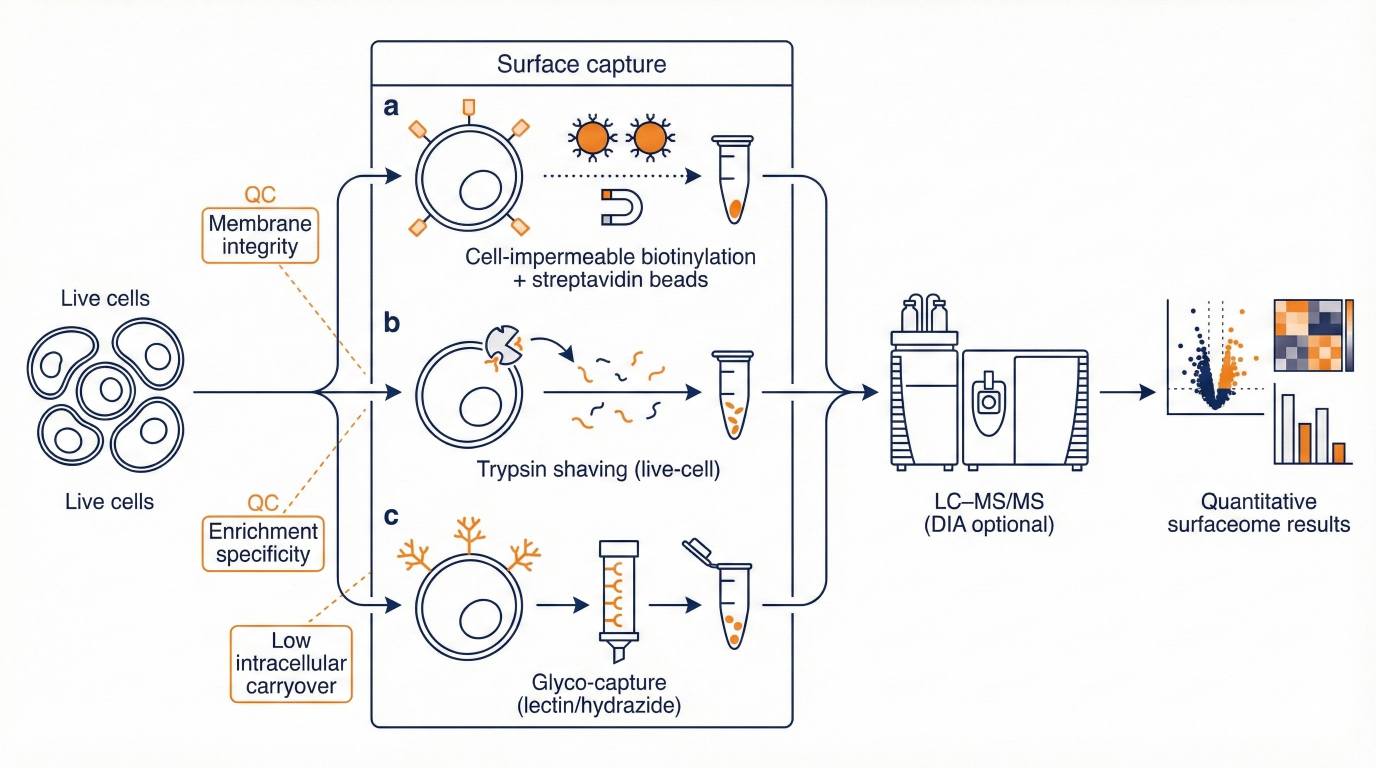
Cell Surface Proteomics Platform (Enrichment Chemistry, LC–MS/MS, DIA)
This service uses a surface-focused enrichment strategy paired with LC–MS/MS. Optional modules can be selected to align with specific biological questions and membrane protein challenges.
Core platform components
- Surface-directed enrichment (cell surface labelling and affinity capture, or surface peptide release)
- Membrane-aware sample preparation to improve recovery of hydrophobic proteins
- LC–MS/MS for identification and quantification
- Optional DIA proteomics for consistent quantitative comparisons
- QC framework using marker-based logic and membrane integrity checks
Technical note for difficult multi-pass targets
For GPCRs and other multi-pass transmembrane proteins, membrane-aware solubilisation options (for example, detergents, nanodiscs, or SMA-based extraction) may be evaluated to support recovery and LC–MS/MS performance.