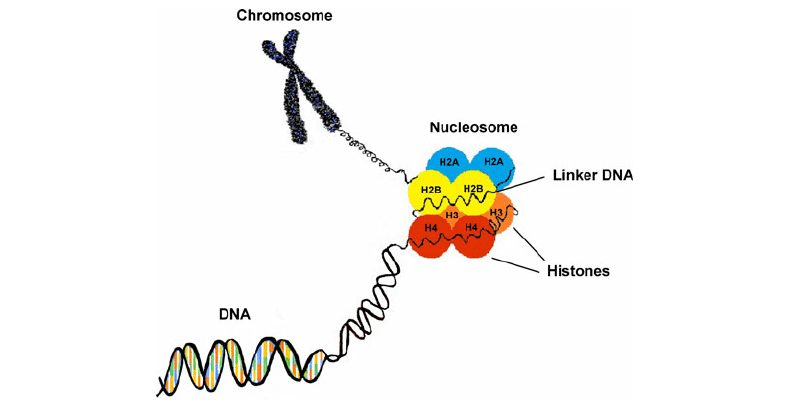
What are histones?
Found in eukaryotic cells, histones are proteins that package DNA into nucleosomes. Histones H2A, H2B, H3, and H4 are defined as core histones that exist as dimers to form octameric nucleosome core, which are wrapped by approximately 146 bp DNA. The nucleosomes are joined by DNA and histone H1 to form chromatin fibers. As essential as they are in chromosome assembly, histones can be post-translational modified to regulate the dynamic expression of genes.
Histone modification
Histone modifications, including methylation, acetylation, phosphorylation, ADP-ribosylation, ubiquitination, and subgroup ubiquitination, begin from the N-terminal tails and move on to the globular domain. These modifications contribute to the binding of other proteins to DNA, resulting in synergistic or antagonistic effects to regulate gene transcription. Take histone acetylation as an example. Histone acetylation and deacetylation are in dynamic equilibrium. The levels of histone acetylation are regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). HATs transfer the acetyl group of CoA and acetyl-CoA to specific lysine residues at the histone N-terminus. After the histone carries the negatively charged acetylated group, it is mutually exclusive with the negatively charged DNA. Therefore, DNA entanglement becomes relaxed and can bind to transcription factors to promote gene expression.
Accurately identifying different modified domains within the same protein or the same modified domain in different proteins can be a challenge. Different types of modifications can combine to regulate the protein functions. Therefore, one study on the histone modification is the relationship between the combinatorial post-translational modification of histones and cellular biological function. Below, we will provide two analytical strategies for histone modification analysis.
Histone post-translational modification analysis strategy
1. Extraction and purification of histones
Hypotonic lysis buffer is added into the cell pellet. The cell membrane is cleaved by mechanical shear force and the nuclei are separated by centrifugation at low speed. The nuclei are then broken down and histones obtained by acid extraction or high salt extraction. HPLC is used to isolate and purify various histone components for further analysis.
2. Mass spectrometry (MS)
A. Middle-down MS
Analyzing histone modification using middle-down MS requires the enzymatic hydrolysis of specific proteases, such as GluC or Asp-N, or target proteins with known sequences to produce large peptide fragments or macromolecular peptides.
B. Top-down MS
The top-down analysis method, often used to analyze purified complete protein, can quickly detect the modified histones in a single experiment without the need for enzymatic hydrolysis. This method facilitates rapid detection of histone modification in a single experiment.
Creative Proteomics provides Q Exactive™ HF Hybrid Quadrupole-Orbitrap™ MS to help you analyze histone modifications.