Author: Creative Proteomics Scientific Team — senior proteomics scientists and bioinformaticians (Creative Proteomics). The team specializes in Orbitrap DIA/TMT phosphoproteomics with IMAC/TiO2 enrichment; see our service overview (Creative Proteomics phosphoproteomics services).
Introduction
Phosphoprotein measurements underpin pathway biology, target validation, and translational biomarker work. Most programs move from broad discovery to targeted validation. This guide compares MS-based phosphoproteomics with immunoassays—traditional ELISA and multiplex platforms (MSD electrochemiluminescence and Luminex xMAP)—to help you choose the right tool at each stage.
We will assess platforms across sensitivity and dynamic range, multiplexing and throughput, sample input, site localization, turnaround, and indicative costs. The goal: fit-to-purpose decisions, not a single winner.
Key takeaways
- MS-based phosphoproteomics maps phosphorylation sites proteome-wide and localizes modifications; it is strongest for discovery and mechanism.
- ELISA, MSD, and Luminex excel in targeted validation with high throughput; they quantify predefined phospho-epitopes but do not provide MS-level site localization.
- DIA/TMT, proper enrichment (IMAC/TiO2), and strict QC (1% PSM/peptide FDR; controlled site FLR) are essential for credible phosphosite data.
- A hybrid pipeline—MS discovery → short list → targeted MS or immunoassay validation—usually yields the most robust, reproducible outcomes.
Platform fundamentals
What MS-based phosphoproteomics measures
MS-based phosphoproteomics enriches phosphopeptides (e.g., IMAC, TiO2), separates by LC, and identifies/quantifies by HRAM MS/MS. It reports site-resolved changes, including positional isomers when fragment evidence permits. DIA-based strategies improve completeness and reproducibility versus stochastic DDA, while TMT multiplexing boosts precision across many conditions. For an overview of global phosphoproteomics DIA strategies and performance, see the approach described in the 2021 Nature Communications study on hybrid spectral libraries and reproducibility in human/yeast systems, presented as an authoritative summary of DIA global phosphoproteomics strategy and performance context (Kitata 2021, Nature Communications). For practical workflow context, see the phosphoproteomics workflow overview on the Creative Proteomics site.
What ELISA detects
ELISA quantifies a single phospho-epitope per assay using phospho-specific antibodies. Readouts are colorimetric or luminescent and kit-validated for range and precision. Ultra-sensitive ELISA-like formats (e.g., ProQuantum) can reach sub–pg/mL for certain targets as indicated by the product page for Human Tau pT181 with sub–pg/mL sensitivity (Thermo Fisher ProQuantum). Classic phospho-ELISAs typically achieve low pg/mL to ng/mL sensitivity depending on target and matrix.
Discovery vs validation contexts
Discovery programs rely on MS-based phosphoproteomics to reveal site-specific pathway dynamics, detect novel sites, and support network modeling when unbiased breadth, site localization, or pathway kinetics are required. Validation programs favor ELISA, MSD, and Luminex to quantify predefined phospho-epitopes at high throughput across large cohorts once targets are known and antibodies are well-qualified.
Quantitative benchmarks
Sensitivity, dynamic range, and multiplexing
- MS-based phosphoproteomics: Practical dynamic range of 4–6 orders is achievable depending on instrument and workflow. DIA strategies improve reproducibility; TMT enhances precision and depth across channels, with comparative studies reporting strong precision for multiplexed Orbitrap analyses as discussed in the Anal Chem 2022 comparative study on quantification methods for tumor proteomes (Zhang et al., 2022).
- ELISA (single-plex): Typical dynamic range spans 2–4 orders (kit-dependent). Sub–pg/mL sensitivity is achievable in specialized formats (e.g., ProQuantum pTau pT181) as noted on the product page (Thermo Fisher ProQuantum Human Tau assay) (Thermo Fisher ProQuantum).
- MSD multiplex ECL: Commonly delivers 4–5 orders of linear dynamic range with multi-spot plates (panel-dependent); kit datasheets provide LLOQ/ULOQ and precision for each analyte on the manufacturer’s library (consult specific panels at the vendor site).
- Luminex xMAP: Multiplex capacity ranges from 50 analytes (MAGPIX) to 500 (FLEXMAP 3D/INTELLIFLEX), with typical dynamic range ≥3.5 logs on MAGPIX and ≥5.5 logs on INTELLIFLEX as stated in the official RUO system specification sheet for INTELLIFLEX and Diasorin product pages for MAGPIX (INTELLIFLEX RUO spec sheet; MAGPIX product page). Sensitivity is kit-dependent.
Throughput, sample input, and turnaround
- MS-based phosphoproteomics: Per-run throughput is determined by gradient length and multiplex strategy. DIA processes one sample per run; TMT batches 10–18 samples with MS3/real-time search strategies on modern Orbitraps. Typical project turnarounds range from 2 to 12+ weeks depending on queue, sample count, and analysis depth.
- ELISA: 96-well kits routinely process 80–96 samples per plate in 1.5–4 hours.
- MSD: 96-well multi-spot plates with 4–10+ analytes per well; typical assay times 2–4 hours per plate (panel-dependent).
- Luminex: 96‑well bead‑based plates; bead‑based, high‑plex workflows support plate‑scale, high‑throughput cytokine and phospho‑epitope panels—see our Luminex cytokine detection service for typical throughput and assay details (Luminex cytokine detection service).
Cost ranges and resource requirements
Indicative 2026 ranges (vary by region, kit, and scope):
- MS-based phosphoproteomics (label-free DIA): approximately $800–$2,000 per sample (includes digestion, phosphopeptide enrichment, LC–MS/MS acquisition, and basic bioinformatics). Costs trend higher for deep fractionation or extensive fraction-level replicates.
- MS-based phosphoproteomics (TMT multiplexed, per-sample after batching): approximately $300–$800 per sample (TMT labels, cleanup/fractionation, enrichment and pooled LC–MS/MS; per-sample cost decreases with larger plexing and batch size).
- ELISA (commodity 96‑well kits): approximately $30–$280 per sample (kit-dependent; based on 96‑well plate economies after standards/controls are accounted for). Ultra‑sensitive or proprietary kits can sit at the upper end.
- MSD (V‑PLEX/U‑PLEX): panel- and analyte-dependent; typical per-plate or per-panel costs are variable—consult vendor or service provider for quotes. Development or custom panels add substantial validation costs.
- Luminex xMAP: panel- and instrument-dependent; per-plate costs vary widely with plex and reagents—consult vendor or provider for exact pricing. Custom high‑plex panels and assay validation materially increase cost.
Note: the ranges above are indicative 2024–2026 estimates; final prices depend on region, sample matrix, depth of fractionation, number of fractions, validation requirements, and whether analysis is performed in an academic core or commercial lab. Obtain written quotes for project budgeting.
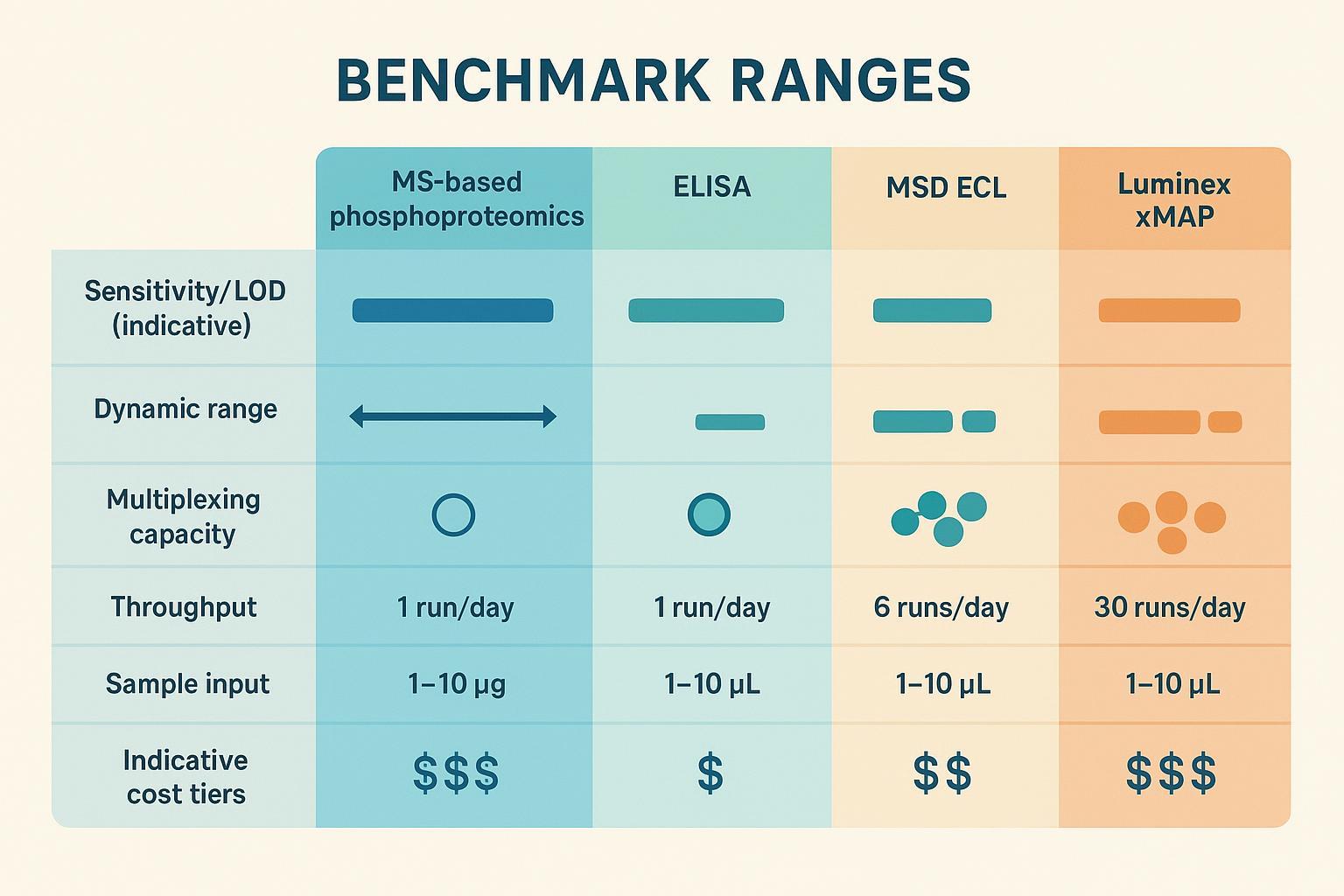
To aid quick scanning, the table below aligns core fields across platforms.
| Platform | What it measures | Typical dynamic range | Multiplex capacity | Sensitivity examples | Sample input | Throughput/turnaround | Site-level resolution |
|---|---|---|---|---|---|---|---|
| MS-based phosphoproteomics | Enriched phosphopeptides identified by LC–MS/MS | ~4–6 orders (workflow-dependent) | TMT 10–18 channels; DIA single-sample per run | Large-scale site detection with DIA/TMT; precision advantages reported for multiplexed Orbitrap analyses (Zhang 2022) | 1–100 µg protein (typical; matrix-dependent) | Runs per day depend on gradient; 2–12+ week projects (queue-dependent) | Yes (localization probabilities; control FLR) |
| ELISA (single-plex) | Single phospho-epitope | 2–4 orders (kit-dependent) | 1 | Sub–pg/mL for select targets (e.g., ProQuantum pTau pT181) (Thermo Fisher ProQuantum) | µL-level lysate/serum per well | 1–4 hours per 96-well plate | No |
| MSD multiplex ECL | Multiple epitopes per well | ~4–5 orders (panel-dependent) | 4–10+ per well | LLOQ/ULOQ in panel datasheets (analyte/matrix-specific) | µL-level per well | 2–4 hours per 96-well plate | No |
| Luminex xMAP | Bead-coded multiplex | ≥3.5 logs (MAGPIX); ≥5.5 logs (INTELLIFLEX) | Up to 50–500 (instrument/panel) | Assay-dependent; instrument enables sub-pg sensitivity with qualified kits (INTELLIFLEX spec) | µL-level per well | High plate throughput; thousands of results/hour possible (MAGPIX page) | No |
QC and site localization
FDR and FLR reporting standards
Community practice is to filter at 1% FDR at PSM and peptide levels and to clearly document methods and thresholds. HUPO/HPP guidelines emphasize rigorous reporting and evidence standards for data deposition and interpretation in proteomics, including multi-peptide evidence for protein-level claims; see the 2024 report on HPP data interpretation guidelines for an overview of documentation standards (Omenn et al., 2024). For phosphosite claims, control of False Localization Rate (FLR) is equally important. Independent evaluations argue for explicit FLR estimation and often recommend controlling global FLR near ~5% for reliable site calls in large datasets; see analyses of localization uncertainty and FLR estimation published in 2022 with discussion of reliable thresholds and multi-evidence validation for phosphosites (Ramsbottom et al., 2022).
Localization probability thresholds and tools
Localization algorithms report probabilities or scores that should be combined with FDR/FLR controls:
- PTMProphet probabilities: thresholds such as ≥0.75 are common in practice, but stringency should be tuned to keep FLR within acceptable bounds and to require replicate/orthogonal evidence where feasible, as discussed in 2023 methodological overviews and analyses on FLR and localization scoring (Zong et al., 2023).
- Ascore: conservative cutoffs around ≥20 are often used; lowering thresholds increases risk of mislocalization according to recent reviews that summarize computational approaches and their trade-offs (Joyce et al., 2023).
- PhosphoRS: probability-based localization; thresholds vary with data quality and should be paired with explicit FLR estimation and replicate evidence.
Enrichment strategies and DIA/TMT advances
IMAC/TiO2 remain the workhorses for phosphopeptide enrichment. DIA-based global phosphoproteomics reduces missingness and improves reproducibility versus DDA, while TMT (with MS3 and real-time search) increases precision across many conditions. For DIA global strategy and performance context, see the 2021 Nature Communications study on hybrid spectral libraries (Kitata 2021). For precision advantages of multiplexed Orbitrap quantification approaches, see the Anal Chem 2022 comparative study of quant methods (Zhang 2022).
Disclosure: Creative Proteomics offers Orbitrap-based DIA/TMT phosphoproteomics with IMAC/TiO2 enrichment and site localization bioinformatics. Learn more in the phosphorylation service overview at Creative Proteomics.
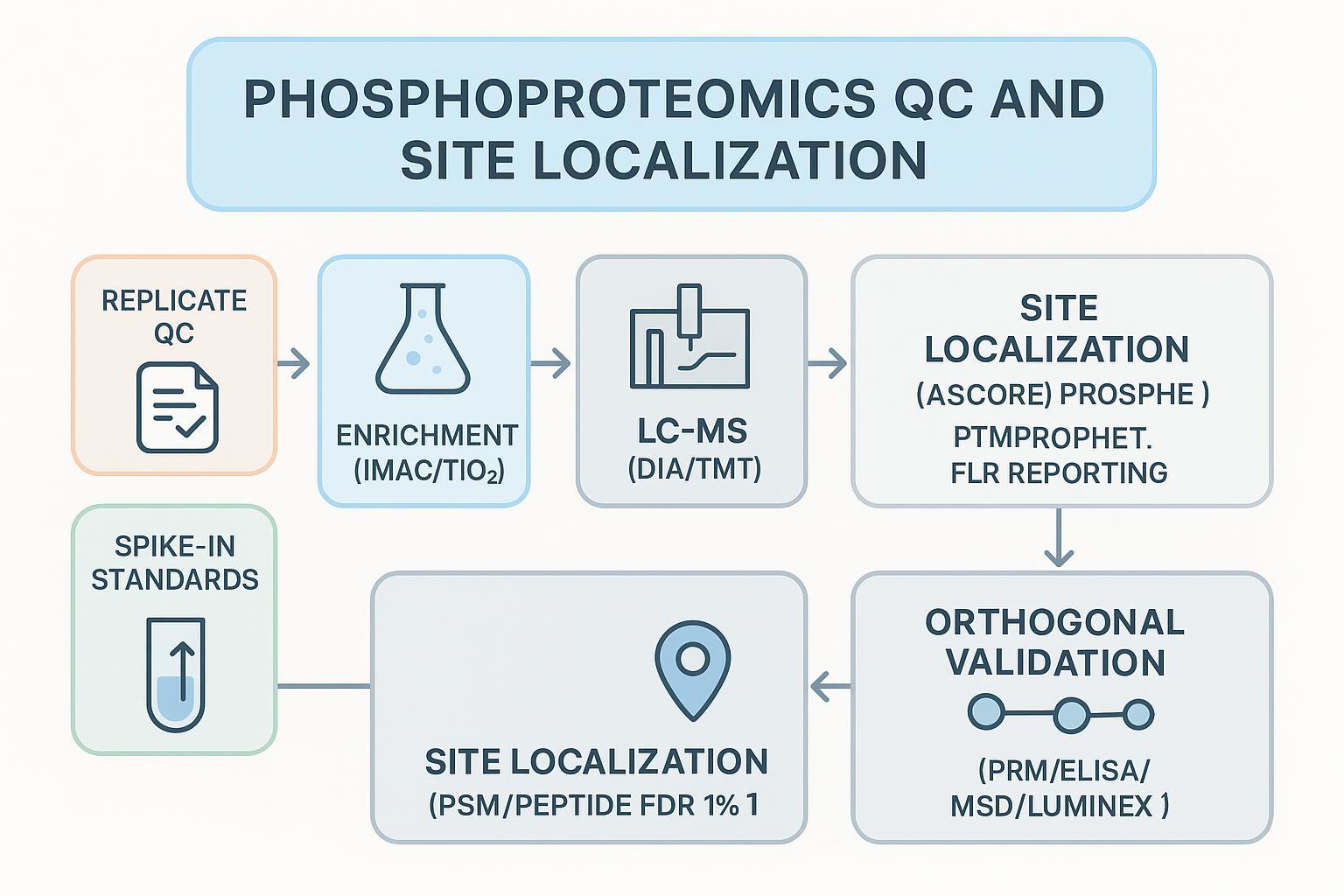
For additional practical guidance on sample prep and analysis tools, see sample preparation best practices on the Creative Proteomics site (Sample prep best practices).
Decision framework and hybrid workflows — how to choose in MS-based phosphoproteomics vs ELISA decisions
Independent case snapshots — peer‑reviewed examples underline typical validation choices. A 2022 global phosphoproteomics study in systemic lupus erythematosus prioritized candidate sites from discovery and confirmed several sites by targeted PRM in patient samples, demonstrating concordant direction and magnitude (see Global Phosphoproteomics Unveils Kinase‑Regulated Networks in SLE, 2022 (PMC9712766)). A 2023 translational review summarizes that targeted MS (PRM/SRM) is the most common orthogonal confirmation for phosphosites and that antibody‑based assays are typically used at the protein/epitope level when validated reagents exist (Mass spectrometry‑based phosphoproteomics in clinical applications, 2023 (PMC10195102)).
When MS-based phosphoproteomics is the better fit: choose it for unbiased discovery, mechanism mapping with site localization, low-stoichiometry signaling, and site-resolved pharmacodynamics. DIA is ideal for completeness and fewer missing values; TMT is ideal when you need multi-condition precision with consistent quantification across many channels.
When ELISA or multiplex immunoassays are optimal: choose ELISA for single-analyte validation when antibodies are well-qualified and you need rapid, economical throughput. Choose MSD for mid-plex panels with wide dynamic range in limited sample volume. Choose Luminex when very high plex (50–500 analytes) and plate-scale throughput are priorities; instrument spec sheets document multiplex capacity and dynamic range (INTELLIFLEX RUO spec).
Hybrid pipeline linking discovery to validation: start with MS discovery (IMAC/TiO2 → LC–MS DIA/TMT) filtered at 1% PSM/peptide FDR; estimate and limit site FLR; require localization probability thresholds and replicate evidence. Form a shortlist by effect size and pathway relevance. Validate with targeted MS (PRM/SRM) when antibodies are unavailable or specificity is uncertain, or with ELISA/MSD/Luminex when qualified antibodies exist and high-throughput cohort testing is needed. Build in spike-in standards, plate controls, blinded replicates, and CV targets (e.g., ≤15% for targeted assays).
If you are scoping a discovery-to-validation plan or need help selecting DIA vs TMT and enrichment strategies for phosphoproteomics, consider a brief consult to pressure-test design, sample requirements, and QC checkpoints through the phosphoproteomics service overview on the Creative Proteomics site for context on services supporting phosphoproteomics studies (Phosphoproteomics service).
Conclusion
There is no universal winner in MS-based phosphoproteomics vs ELISA decisions. MS offers site-resolved discovery power and pathway insight; ELISA, MSD, and Luminex deliver scalable, targeted quantitation. Match the platform to the question, matrix, and sample budget. Run pilots, document FDR/FLR and localization thresholds, and include controls to ensure reproducibility.
For complex studies linking discovery to validation, a hybrid pipeline usually performs best. If you want a second set of eyes on study design, enrichment choice, or targeted follow-up options, you can consult the phosphoproteomics team via the Creative Proteomics service page linked above to discuss a pilot and QC plan.
Our products and services are for research use only.


.jpg)

.jpg)