Introduction to Pan PTM Proteomics
Traditional PTM studies usually focus on a single modification type, such as phosphorylation or glycosylation. While useful, these approaches miss the broader regulatory complexity where multiple PTMs act together to influence protein activity and cellular pathways.
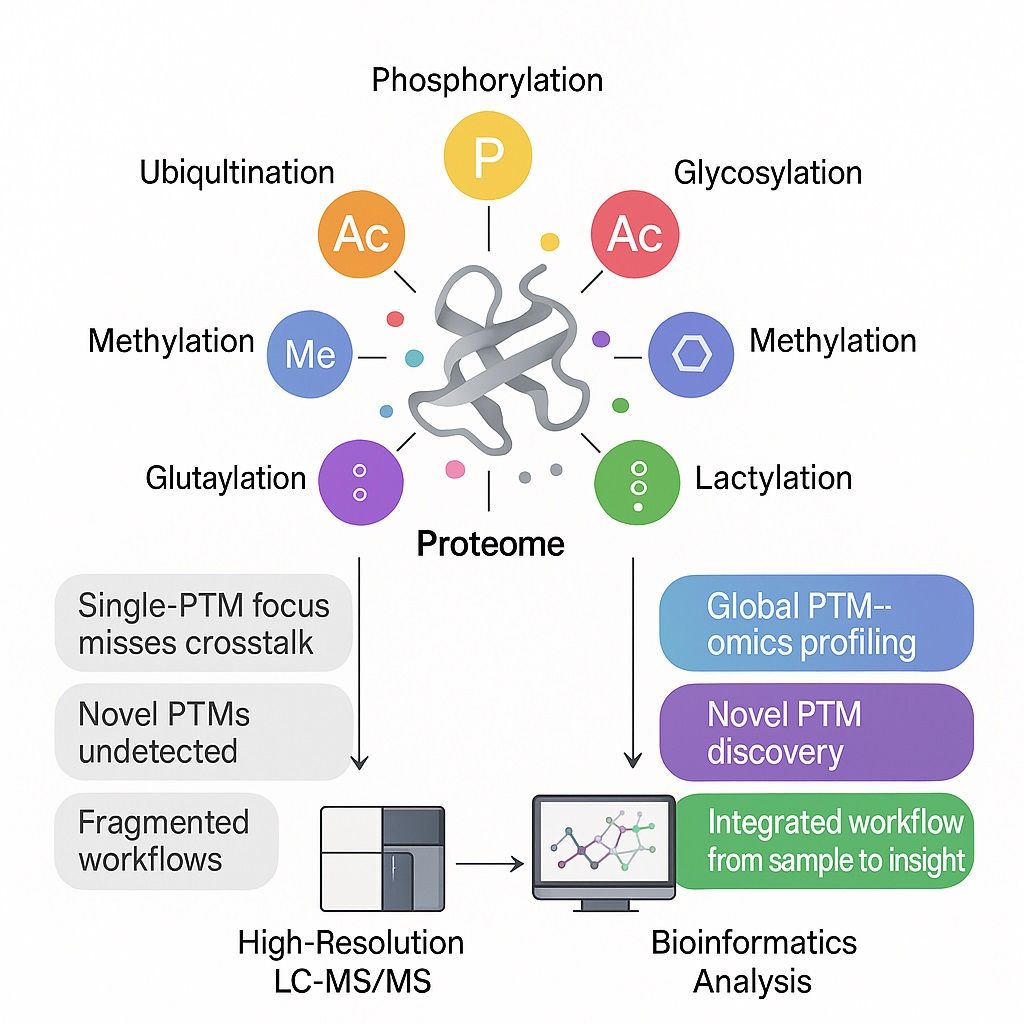
Pan PTM proteomics offers a global post-translational modification (PTM) omics strategy. By analysing a wide range of PTMs simultaneously, this approach enables researchers to achieve proteome-wide post-translational modification analysis in a single workflow.
For scientists working in drug discovery, biomarker development, or systems biology, this comprehensive view provides actionable insights that conventional methods cannot capture. By combining high-resolution mass spectrometry with advanced bioinformatics, Pan PTM proteomics delivers the complete PTM landscape, including both well-characterised and novel modification types.
Why Pan PTM Matters
Traditional proteomics often isolates one post-translational modification (PTM) at a time. This narrow view cannot explain how proteins are regulated in complex biological systems. Many diseases—including cancer, neurodegeneration, and metabolic disorders—arise from the combined effects of multiple PTMs acting together.
Pan PTM proteomics provides a solution. By enabling proteome-wide post-translational modification profiling, this approach delivers:
- Comprehensive coverage: Simultaneous mapping of phosphorylation, glycosylation, acetylation, ubiquitination, methylation, and emerging PTMs.
- Novel PTM discovery: Open-search pipelines detect unexpected modifications that targeted assays often miss.
- Crosstalk insights: Multi-PTM data integration reveals how modifications cooperate or compete on proteins and pathways.
- Translational value: Identification of modification signatures linked to biomarkers, disease mechanisms, or druggable targets.
- Efficiency: One workflow generates a multi-dimensional dataset, reducing the need for multiple experiments.
For researchers focused on specific regulatory layers, Pan PTM can also serve as a discovery-first approach. Results can guide targeted follow-up studies, such as Phosphoproteomics Analysis or Glycoproteomics Analysis.
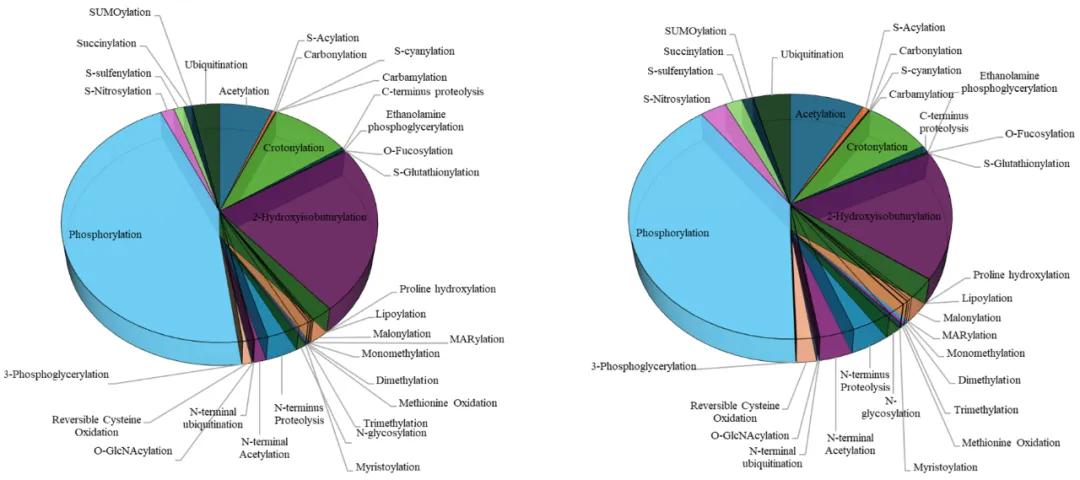
Types of PTMs Analyzed in Pan PTM Profiling
The strength of Pan PTM proteomics lies in its ability to profile a broad spectrum of post-translational modifications in one workflow. Instead of limiting analysis to a single modification type, this approach detects both well-characterized PTMs and emerging regulatory marks.
Common PTMs covered include:
Phosphorylation – key regulator of signal transduction and enzyme activity.
Ubiquitination and SUMOylation – central to protein degradation, DNA repair, and stress response.
Acetylation and acylations (butyrylation, crotonylation, propionylation) – regulators of chromatin state and metabolism.
Methylation (mono-, di-, tri-) – controls transcriptional regulation and protein interactions.
Glycosylation (N-linked, O-linked) – critical for protein folding, stability, and immune recognition.
Lactylation, Succinylation, Malonylation, Glutarylation – newly identified modifications linked to energy metabolism and disease pathways.
Oxidation and Deamidation – spontaneous PTMs associated with protein aging and stress conditions.
This breadth makes Pan PTM an effective discovery platform for proteome-wide post-translational modification analysis. It not only profiles known sites but also uncovers novel modifications that may represent unexplored regulatory mechanisms or therapeutic targets.
Comparison: Pan PTM vs. Conventional PTM Omics
While these targeted approaches are valuable for specific pathways, they miss the bigger picture of protein regulation where multiple PTMs act in concert.
Pan PTM proteomics addresses this limitation by providing multi-PTM profiling in one workflow. This enables both discovery of novel modifications and crosstalk analysis that single-PTM studies cannot achieve.
| Feature |
Pan PTM Proteomics |
Single-PTM Omics (e.g., Phosphoproteomics, Glycoproteomics) |
| Scope |
Global, multi-PTM profiling in one analysis |
Focused on one PTM class per study |
| PTM Types Covered |
10+ (phosphorylation, acetylation, ubiquitination, glycosylation, methylation, lactylation, succinylation, etc.) |
One PTM type only |
| Novel PTM Discovery |
Yes – open-search pipelines detect unexpected modifications |
Rare – relies on targeted enrichment methods |
| Crosstalk Analysis |
Reveals how PTMs cooperate or compete on proteins/pathways |
Usually not captured |
| Applications |
Biomarker discovery, drug target identification, systems biology |
Focused mechanistic insights on one regulatory pathway |
| Cost & Efficiency |
One integrated workflow, reduced redundancy |
Multiple experiments required for different PTMs |
When to Choose Each Approach
- Use Pan PTM proteomics when the goal is comprehensive discovery, biomarker mapping, or hypothesis generation in complex systems.
- Choose targeted PTM omics when you need in-depth coverage of a specific pathway.
Both approaches are complementary: Pan PTM defines the broad landscape, and targeted PTM studies validate and refine the mechanistic insights.
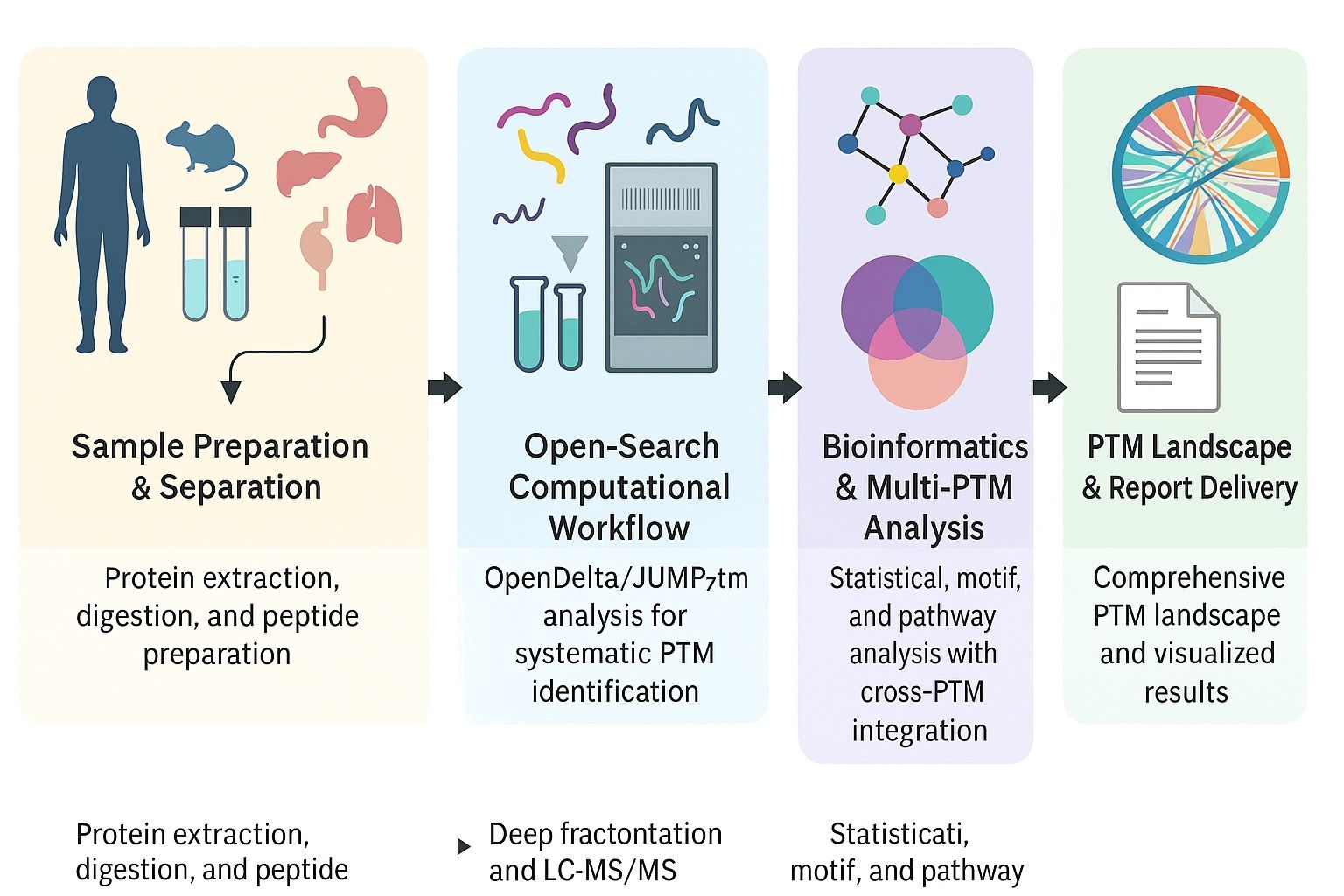
Our Pan PTM Proteomics Workflow
Sample Preparation & Quality Control
- Protein extraction from tissues, cells, or biofluids.
- Protein concentration measurement and SDS-PAGE validation to ensure sample quality.
Enzymatic Digestion & Fractionation
- Proteins digested into peptides using trypsin.
- Desalting and ultra-deep peptide fractionation to improve PTM coverage.
High-Resolution Mass Spectrometry
- LC-MS/MS detects peptides with and without PTMs.
- Ultra-high resolution ensures sensitivity and precise mass shift identification.
Open-Search & Targeted Identification
- Use of advanced pipelines (e.g., OpenDelta, JUMPptm) for systematic PTM discovery and quantification.
- Simultaneous detection of known PTMs and novel, unexpected modifications.
Bioinformatics Analysis
- Statistical QC and clustering (PCA, hierarchical).
- Functional enrichment: Gene Ontology (GO) and KEGG pathways.
- Motif and sequence analysis to define PTM consensus patterns.
- Multi-PTM crosstalk analysis and kinase-substrate enrichment (KSEA).
Reporting & Biological Interpretation
- Delivery of a comprehensive PTM landscape.
- Highlighted candidate proteins and modification sites for biomarker or target validation.
- Data visualizations and actionable insights for research decision-making.
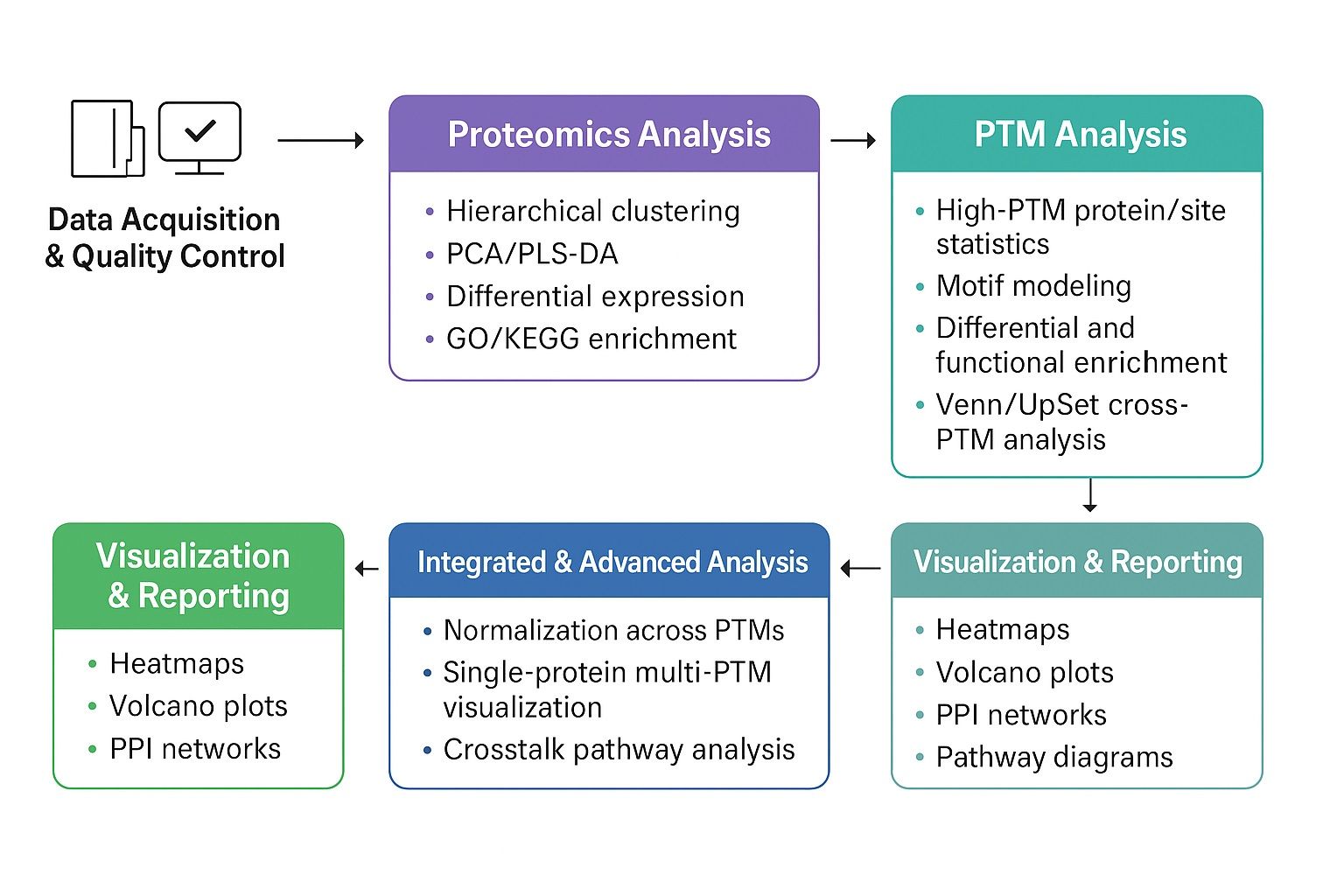
Data Analysis Modules in Pan PTM Proteomics
| Analysis Module |
Specific Analyses |
Value and Application Context |
| Quality Control |
Raw spectra quality assessment, noise filtering, reproducibility checks |
Ensures data reliability and reduces false positives |
| PTM Feature Modeling |
PTM site preference modeling, upstream/downstream amino acid motifs, motif visualization |
Reveals modification rules and supports enzyme mechanism studies |
| Pathway & Network Analysis |
GO/KEGG enrichment, protein–protein interaction (PPI) networks, hub protein identification |
Understands how PTMs regulate biological processes at the systems level |
| Dynamic Regulation |
Time-series or treatment-group PTM dynamics |
Identifies how modifications change in response to stimuli or drugs |
| Cross-PTM Analysis |
Venn/UpSet overlap analysis, synergistic or antagonistic PTM pathway mapping |
Highlights cooperation or competition among different modifications |
| Functional Prediction |
Integration with transcriptomics/metabolomics, kinase-substrate enrichment analysis (KSEA) |
Links PTM changes to gene expression and metabolic pathways |
| Visualization & Reporting |
Heatmaps, volcano plots, network graphs, pathway diagrams |
Provides intuitive interpretation and publication-ready outputs |
Applications of Pan PTM Proteomics
Biomarker Discovery
Identify PTM signatures linked to cancer, metabolic disease, or neurodegeneration. Pan PTM profiling reveals modification sites that correlate with disease progression or therapeutic response.
Drug Target Identification
Detect PTM-regulated proteins and residues that serve as candidate drug targets. This enables proteome-wide post-translational modification analysis to guide therapeutic development.
Mechanistic Studies
Uncover how phosphorylation, acetylation, ubiquitination, and novel PTMs cooperate to regulate signaling, immune response, or stress adaptation.
Aging and Longevity Research
Studies such as pan-PTM profiling in Drosophila have linked modifications to longevity and preserved muscle function, offering models for human healthspan research.
Systems Biology
Map PTM networks and crosstalk to understand molecular complexity at the systems level. These insights support biomarker validation.
Why Choose Creative Proteomics
Selecting the right partner for Pan PTM proteomics is not only about instrumentation—it is about expertise, workflow reliability, and the ability to translate data into insight. Creative Proteomics differentiates itself through:
Proven Track Record in PTM-Omics
Our team has delivered projects spanning phosphorylation, glycosylation, acetylation, and ubiquitination for global pharma and academic clients. Pan PTM builds on this foundation by integrating multiple PTM types into one comprehensive service.
Advanced Computational Pipelines
Beyond standard search engines, we deploy open-search workflows (e.g., OpenDelta, JUMPptm) tailored for PTM discovery. This enables detection of rare or novel modifications that traditional proteomics pipelines often overlook.
Depth and Sensitivity at Scale
Our LC-MS/MS systems are optimized for ultra-deep peptide fractionation, allowing detection of thousands of PTM sites in a single study—even in low-abundance proteins or challenging sample types.
Integrated Bioinformatics & Interpretation
We don’t stop at raw data. Our bioinformatics team delivers network-level insights, pathway annotations, and prioritized PTM sites, ensuring data can directly inform biomarker or target validation strategies.
Cross-Industry Experience
We support biopharma R&D pipelines, academic discovery research, and CRO collaborations, adapting reporting formats and project management to client-specific needs.
FAQs
What is Pan PTM proteomics and how does it differ from traditional PTM profiling?
Pan PTM proteomics is a global, multi-modification approach that concurrently profiles many post-translational modification types (phosphorylation, acetylation, ubiquitination, glycosylation, methylation, lactylation, oxidation, etc.) across the proteome, allowing discovery of novel modifications and cross-PTM crosstalk that typical single-PTM workflows cannot capture.
Can you identify novel or unexpected PTMs, not only known ones?
Yes — by leveraging open-search and discovery pipelines (e.g. OpenDelta, JUMPptm), our platform can detect both canonical and previously unreported modifications, expanding the regulatory landscape beyond what targeted workflows allow.
Do I need enrichment steps for Pan PTM proteomics?
It depends on the sample complexity and the abundance of modifications of interest; enrichment is often helpful to boost sensitivity for low-abundance PTMs, but our deep fractionation and open-search methods mitigate this need in many use cases.
What sample types are accepted and what are input requirements?
We accept diverse sample types (cells, tissues, biofluids). For best results, samples should be properly lysed, reduced, alkylated, and free of interfering substances; we provide detailed guidelines to ensure compatibility with our workflow.
How can Pan PTM proteomics support biomarker discovery and drug target research?
By generating a multi-dimensional PTM landscape, our service highlights modification sites that correlate with disease phenotypes or treatment responses, enabling you to prioritize candidates for downstream validation or functional studies.
Is Pan PTM proteomics compatible with downstream targeted PTM studies?
Absolutely — Pan PTM profiling offers a discovery layer, and you can follow up with more focused methods such as phosphoproteomics, glycoproteomics, acetylomics or ubiquitylomics to validate and quantitate specific PTM sites.
Reference
- Poudel, S., Chuang, CL., Shrestha, H.K. et al. Pan-PTM profiling identifies post-translational modifications associated with exceptional longevity and preservation of skeletal muscle function in Drosophila. npj Aging 11, 23 (2025).
- Siemes, D., Voss, H., Benvenuti, F. et al. A reference database enabling in-depth proteome and PTM analysis of mouse immune cells. Sci Data 12, 596 (2025).