Introduction
Precision Medicine is designed to adjust the prevention and treatment of diseases according to individual differences in each patient. To achieve this goal, accurate diagnosis is required. Changes in the human body at the molecular level (such as proteins, RNA, DNA, and metabolic intermediates) contain plenty of potential diagnostic messages and therapeutic targets. Collecting more molecular evidence through omics (e.g., proteome, genome, metabolome) can be based on a comprehensive reflection of different molecular evidence to more accurately judge and develop treatment for diseases. Transition from a single biomarker to multiple biomarkers (i.e., molecular characteristics) in near future is expected to significantly improve the accuracy of the diagnosis, and the combination therapy with multiple targeted drugs will become more common. In addition to early diagnosis, biomarkers can provide doctors more information, such as the optimal treatment options based on biomarker evidence (predictive biomarkers), and prognostic judgments of disease progression monitoring (prognostic biomarkers). The problem is how can we connect clinical disease with the abundant information contained in clinical samples? And how to adopt effective technologies for screening?
In order to better screen Biomarker and therapeutic targets, the study needs to come up with well-defined clinical scientific questions, such as how to distinguish diseased and healthy people; how to more carefully distinguish the specific stage in the occurrence, development and treatment of diseases.
Case Study
In the resolution of clinical problems, large-scale clinical samples are applied to the screening progress, and the sample size usually increases with the sufficiency of the evidence required by the research. Below, we select a article published in high-scoring academic journals, highly recognized by international peers, to analyze how to use precious clinical samples to conduct research from the protein level. The analysis mainly includes four aspects:
- Propose clear scientific questions
- How to select samples for scientific issue
- The techniques adopted
- Study results
- Case: Proteomic Architecture of Human Coronary and Aortic Atherosclerosis

1.Propose clear scientific questions
At the molecular level, atherosclerosis is thought to be a pathological feature formed by the combination of hundreds of proteins inside and outside the cell to alter the cytobiology process and the vascular local environment. The inability to perform individualized treatment through the diagnosis of early atherosclerosis severely hampers the control and treatment of coronary heart disease. In order to improve early disease detection and delay, even interrupt the disease progression before clinical symptoms occur, it is necessary to recognize the dynamic variations and characteristic patterns of arterial protein networks. A comprehensive understanding of arterial protein networks and their changes in early atherosclerosis can screen for new biomarkers for disease detection and a better target for treatment.
2. How to select samples for scientific issues
100 pairs of autopsy specimens within 24 hours of death, 1 g of each of the abdominal aorta (AA) and left anterior descending (LAD) coronary arteries. The pathologists grade the atherosclerotic features of the 10 mm long distal abdominal aorta and the 5 mm long LAD coronary intima surface: (1) fatty streaks (FS); (2) fibers plaques (FPs); (3) complex lesions; (4) calcified lesions, followed by proteomic analysis.
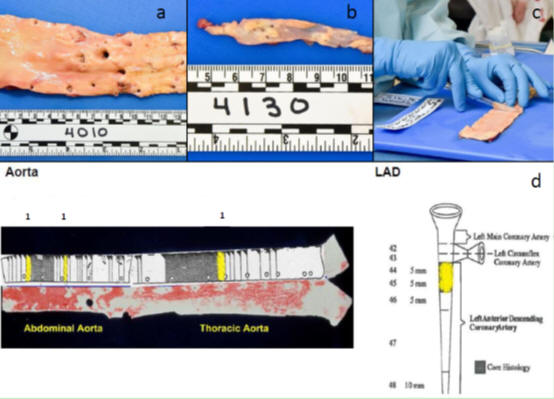
3. The techniques adopted
Label-free proteomics preliminary screening and identification
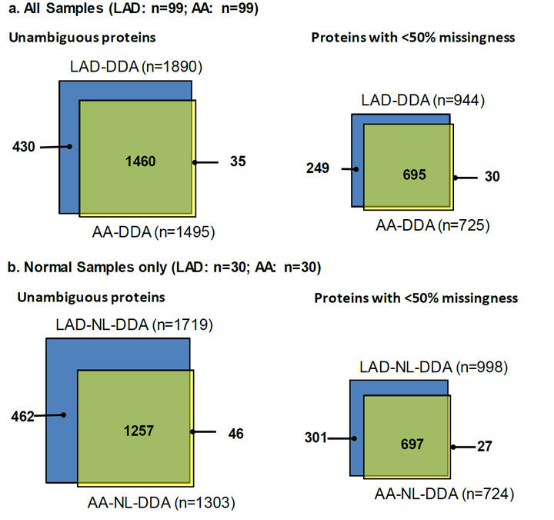
DIA (Data independent acquisition, also known as SWATH) with higher sensitivity and accuracy for quantitative verification. The changes in protein content of FP (i.e. fibrous plaque) vs NL (i.e. normal) samples of LAD and AA were compared respectively. Sample size LAD (FP n=15; NL n=30) and AA (FP n=9; NL n=18)
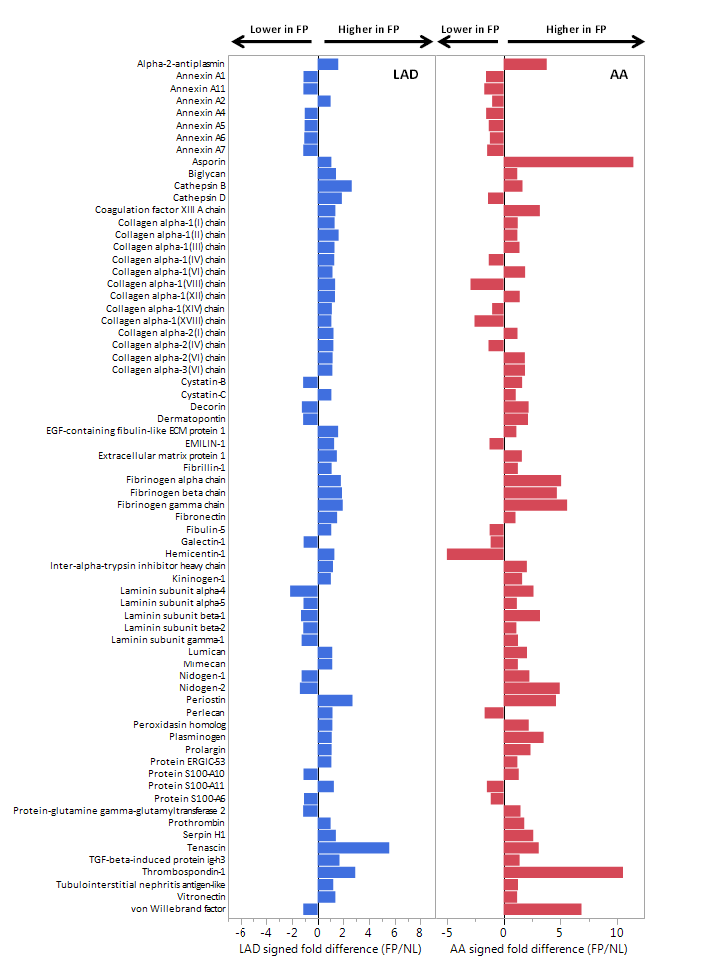
4. Study Results
After the proteomic screening by vascular tissues, differential proteins with secreted potential were detected in serum samples. Comparing the fasting serum protein levels of 45 women with angiographically confirmed coronary atherosclerosis (cases) and 41 women of similar age without coronary artery disease (control group), 15 layers of serum protein were found to be able to predict early atherosclerosis, AUC = 0.93, CI = 95%.
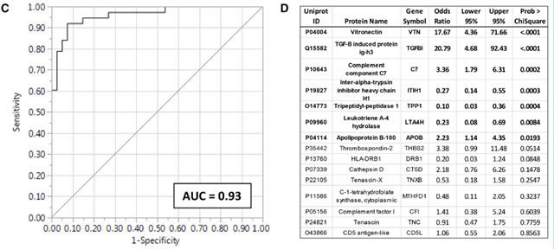
Reference:
- Herrington, David M., et al. "Proteomic Architecture of Human Coronary and Aortic Atherosclerosis." Circulation 137.25 (2018): 2741-2756.
