SILAC-based CoIP-MS Analysis
Why Quantitative Protein Interaction Analysis Matters?
Protein-protein interactions (PPIs) are dynamic and context-dependent. Changes in interaction strength or composition often reflect pathway activation, disease states, or drug-induced perturbations. Traditional qualitative approaches frequently fail to distinguish specific interactors from nonspecific background proteins, limiting biological interpretation.
Quantitative proteomics-based PPI analysis addresses this limitation by enabling direct comparison between experimental and control conditions. In drug discovery, this is particularly crucial for target validation, mechanism-of-action studies, and profiling of off-target interactions. SILAC-based CoIP-MS has become a preferred strategy for high-confidence interaction analysis in complex biological systems.
What Is SILAC-based CoIP-MS?
Stable Isotope Labeling by Amino acids in Cell culture (SILAC) is a metabolic labelling technique in which cells incorporate isotopically labelled amino acids during protein synthesis. When paired with co-immunoprecipitation (CoIP) and LC-MS/MS, SILAC enables accurate relative quantification of proteins co-enriched with a target bait protein.
In a typical SILAC-based CoIP-MS experiment, experimental and control cell populations are differentially labelled with "heavy" and "light" amino acids. After immunoprecipitation of the protein of interest, samples are mixed early in the workflow and analysed together by mass spectrometry. The resulting heavy-to-light peptide ratios enable a direct, internal comparison of protein abundance, allowing for confident discrimination between specific interactors and nonspecific binders. This approach is widely recognised as a benchmark method for quantitative interactomics.
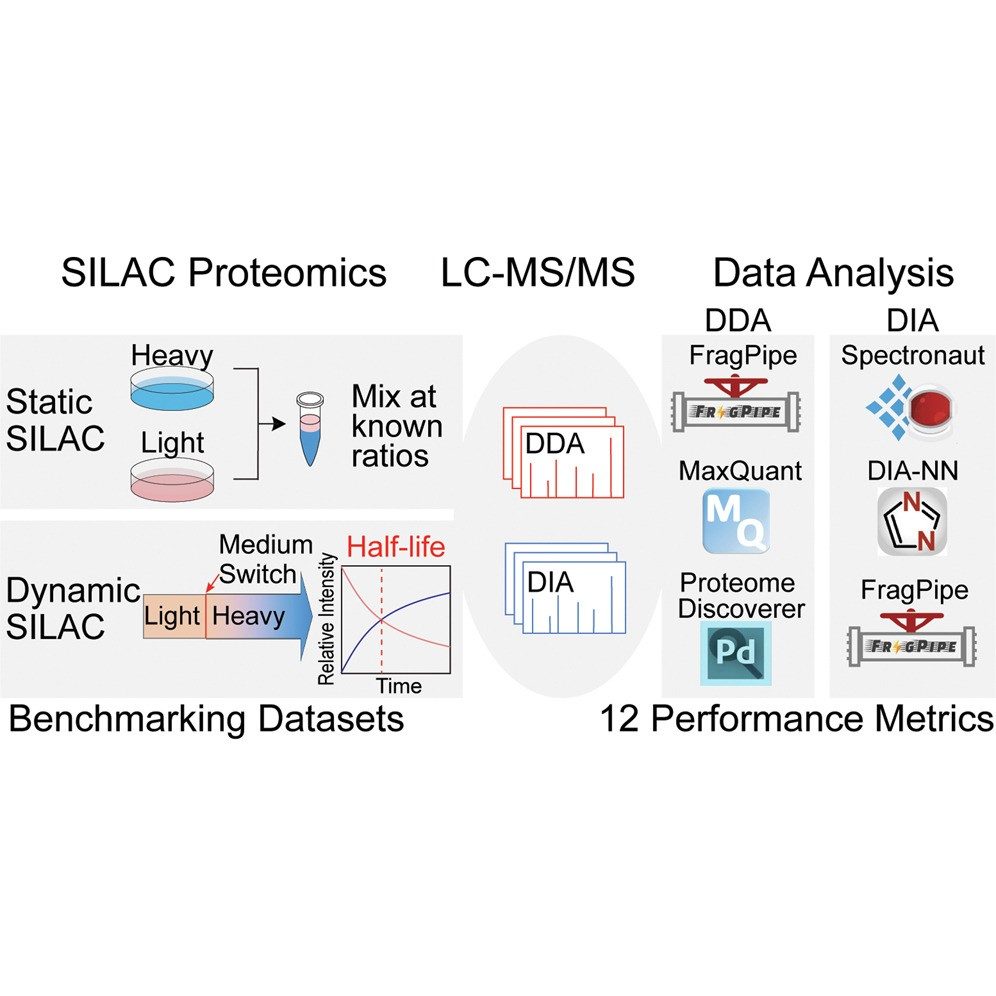
Figure 1. SILAC proteomics workflows and data analysis platforms (Frankenfield A M, et al. 2025).
Advantages of SILAC-based Proteomics
- In vivo labelling: SILAC is ideal for studies that analyse protein interactions and changes in the natural environment.
- High labelling efficiency and stability: SILAC can label almost all proteins in the cell, typically up to 99%.
- High sensitivity: SILAC can detect and quantify low-abundance proteins and is suitable for the identification of low-expressed proteins.
- Dynamic proteome analysis: SILAC is well-suited to studying the dynamics of the proteome under different conditions or time points.
Why Combine SILAC with CoIP-MS?
The integration of SILAC with CoIP-MS offers several critical methodological advantages over conventional IP-MS or post-lysis chemical labelling strategies:
- Reduced Experimental Bias: Isotope labelling occurs during cell growth, prior to protein extraction, minimising variability introduced by sample handling and processing.
- Early Sample Mixing: Heavy and light samples are combined immediately after immunoprecipitation, ensuring identical downstream processing and high quantitative precision.
- Physiological Relevance: Protein interactions are captured in living cells, preserving native conformations and post-translational modifications.
- Low False-Positive Rates: Quantitative comparison against controls enables statistical filtering of nonspecific background proteins.
Comparison with Other Quantitative Proteomics Strategies
TMT (Tandem Mass Tag) labeling enables the simultaneous comparison of multiple samples, making it ideal for large-scale studies or experiments that require high-throughput analysis. Unlike SILAC, TMT labels proteins after digestion, which can sometimes introduce variability in quantification; however, it works well with samples that cannot be metabolically labelled, such as primary tissues.
iTRAQ (Isobaric Tags for Relative and Absolute Quantitation) functions similarly to TMT, chemically tagging peptides post-digestion for simultaneous comparison across samples. iTRAQ is useful for analyzing multiple conditions or treatments in a single experiment, but can be more sensitive to technical variability in sample preparation. SILAC introduces the label during cell growth, so heavy and light samples are mixed early in the process. This reduces experimental error, preserves native protein states, and provides more reliable relative quantification, especially for detecting subtle changes in protein interactions.
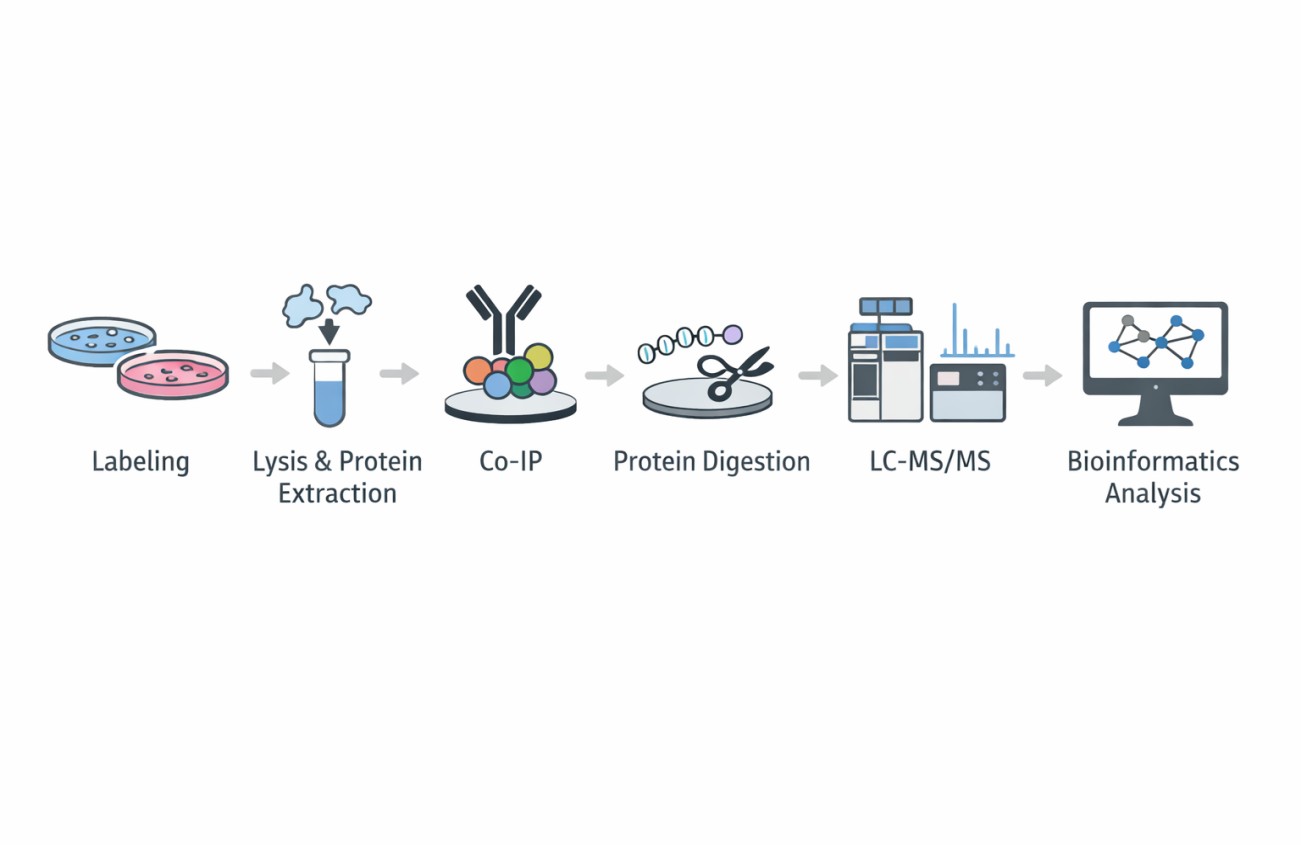
Experimental Workflow of SILAC-based CoIP-MS
- SILAC Cell Culture and Labelling: Cells are cultured in defined media containing heavy or light amino acids until complete isotope incorporation is achieved.
- Cell Lysis and Protein Extraction: Optimised lysis conditions are applied to preserve protein complexes and post-translational modifications.
- Antibody-based Co-IP: Target proteins and their interacting partners are enriched using validated, high-specificity antibodies.
- Efficiency Evaluation and Sample Mixing: IP efficiency is assessed, followed by early mixing of labelled samples.
- Protein Digestion and Peptide Separation: Proteins are enzymatically digested and peptides are separated using nano-liquid chromatography.
- High-resolution LC-MS/MS Analysis: Peptides are analysed on advanced Orbitrap platforms to achieve high mass accuracy and depth of coverage.
- Quantitative Data Processing and Bioinformatics Analysis: Interaction candidates are identified, quantified, statistically filtered, and functionally annotated.
Data Output and Bioinformatics Interpretation
- Detailed experimental procedures and QC documentation
- Mass spectrometry acquisition parameters
- Protein identification and quantitative ratio tables
- Statistical filtering of interaction candidates
- Interaction network visualization and functional annotation
- Complete raw MS data files
Applications in Biomedical and Pharmaceutical Research
SILAC-based CoIP-MS is widely applied across multiple research areas, including:
- Signal transduction and pathway mapping
- Drug target identification and validation
- Mechanism-of-action studies for small molecules and biologics
- Comparative interactomics under disease or treatment conditions
- Functional characterization of protein complexes
Sample Requirements
| Sample type | Actively growing, SILAC-compatible cultured cells |
| Cell number | ≥ 5 × 10⁶ cells per condition |
| Protein amount | ≥ 200–300 μg total protein per IP |
| Controls | Matched negative control (e.g., IgG or empty vector) |
| Replicates | At least two biological replicates |
Why Choose Creative Proteomics for SILAC-based CoIP-MS?
- State-of-the-art Orbitrap mass spectrometry platforms
- Experienced proteomics scientists and bioinformatics experts
- Rigorous QC and standardized workflows
- End-to-end project management from experimental design to data interpretation
FAQ
-
Q1: How does antibody specificity influence data quality in SILAC-based CoIP-MS?
A1: Antibody specificity is a major determinant of experimental success. Poorly characterized or low-affinity antibodies can lead to inefficient enrichment or high background binding, compromising quantitative interpretation. Antibody validation, use of appropriate controls (e.g., IgG pull-downs), and Optimisation of binding conditions are essential to ensure reliable interaction data.
-
Q2: When should SILAC-based CoIP-MS be preferred over label-free IP-MS?
A2: SILAC-based CoIP-MS is preferred when high quantitative precision and low false-positive rates are required, particularly for comparative studies. Label-free IP-MS may be suitable for exploratory analyses or sample-limited systems, but generally exhibits higher variability and reduced quantitative confidence compared with SILAC-based approaches.
-
Q3: Can SILAC-based CoIP-MS be used for time-course or dynamic interaction studies?
A3: Yes. SILAC-based CoIP-MS is well-suited for time-course experiments, as it enables the comparison of interaction profiles across different time points following stimulation or drug treatment. When combined with experimental designs such as pulse-SILAC or label-swap strategies, it allows quantitative tracking of dynamic changes in protein complex assembly and disassembly.
-
Q4: What is the typical project timeline for a SILAC-based CoIP-MS study?
A4: Project timelines depend on cell growth rates, labelling durations, and experimental complexities. In general, SILAC labelling and cell expansion may require several cell doublings, followed by immunoprecipitation, MS analysis, and bioinformatics processing.
Demo
Demo: SILAC–based quantitative MS approach for real-time recording protein-mediated cell-cell interactions
This study extended SILAC-based quantitative mass spectrometry to monitor changes in protein secretion in a tumor-microenvironment co-culture system. While focusing on secreted proteins, the approach demonstrates how SILAC labelling coupled with LC-MS/MS can quantitatively compare protein levels between complex biological conditions, a principle fundamental to SILAC-CoIP comparative PPI analyses.
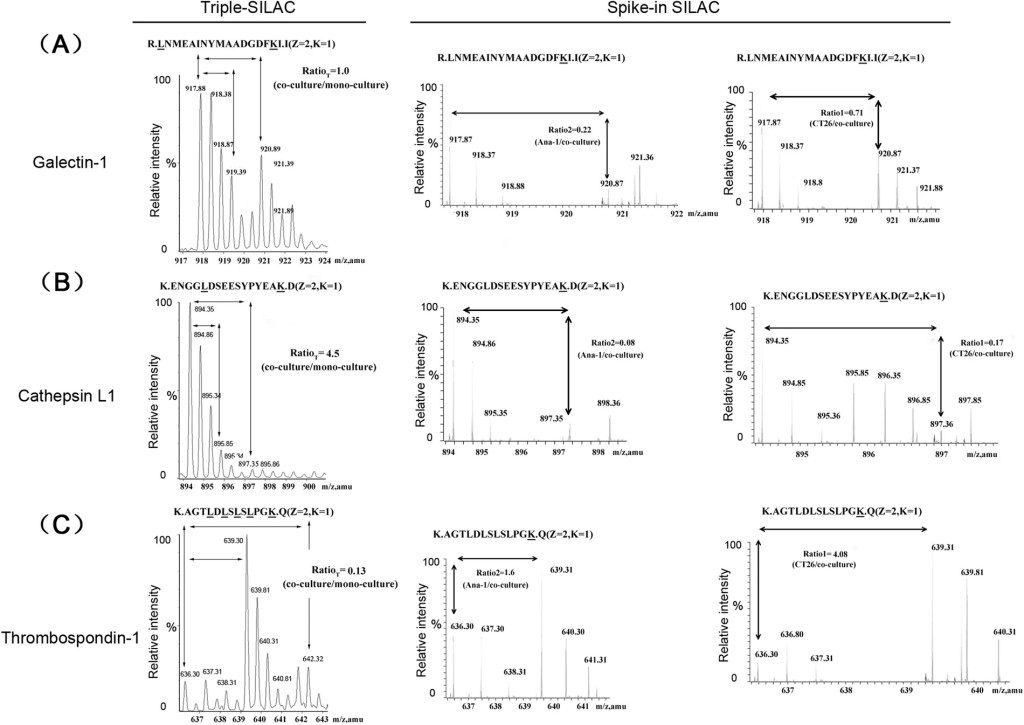
Figure 2. Quantitative MS peaks of three proteins identified by spike-in SILAC and Triple-SILAC in mouse Ana-1 and CT26 co-culture system (Wang X, et al., 2018).
-
Case Study
Case: Triple SILAC Identified Progestin-Independent and Dependent PRA and PRB Interacting Partners in Breast Cancer
Background
Progesterone receptor (PR) signalling plays a central role in breast cancer biology. Two major PR isoforms — PRA and PRB — have overlapping yet distinct biological functions. Understanding the differential protein interactions of these isoforms is critical to elucidating isoform-specific mechanisms in hormone responses and breast cancer progression.
Purpose
To systematically identify and compare protein complexes interacting with PRA and PRB under both progestin-independent and progestin-dependent conditions using a quantitative SILAC-based proteomics approach. The goal was to generate comprehensive interactome maps for each receptor isoform that reflect functional differences.
Methods
- Triple SILAC labelling was employed, where cells expressing HA-tagged PRA or PRB were labelled with distinct stable isotope media (light, medium, heavy), enabling robust quantitative comparisons across conditions.
- Affinity purification (Co-IP) of PRA or PRB complexes was performed using the HA tag.
- Peptides were analysed by LC-MS/MS, and quantitative comparisons of heavy/light SILAC ratios distinguished specific interacting proteins for each receptor isoform and ligand state.
- Both forward and reverse SILAC experiments were applied to minimise labelling bias and experimental noise.
Results
- The study produced differential interaction profiles for PRA and PRB, identifying common and unique interacting partners for each isoform.
- Multiple proteins involved in transcriptional regulation, chromatin remodeling, and nuclear signalling were reproducibly detected as isoform-specific interactors.
- Progestin‐dependent interactions were also identified, revealing how ligand binding modulates receptor complexes differently for PRA and PRB.
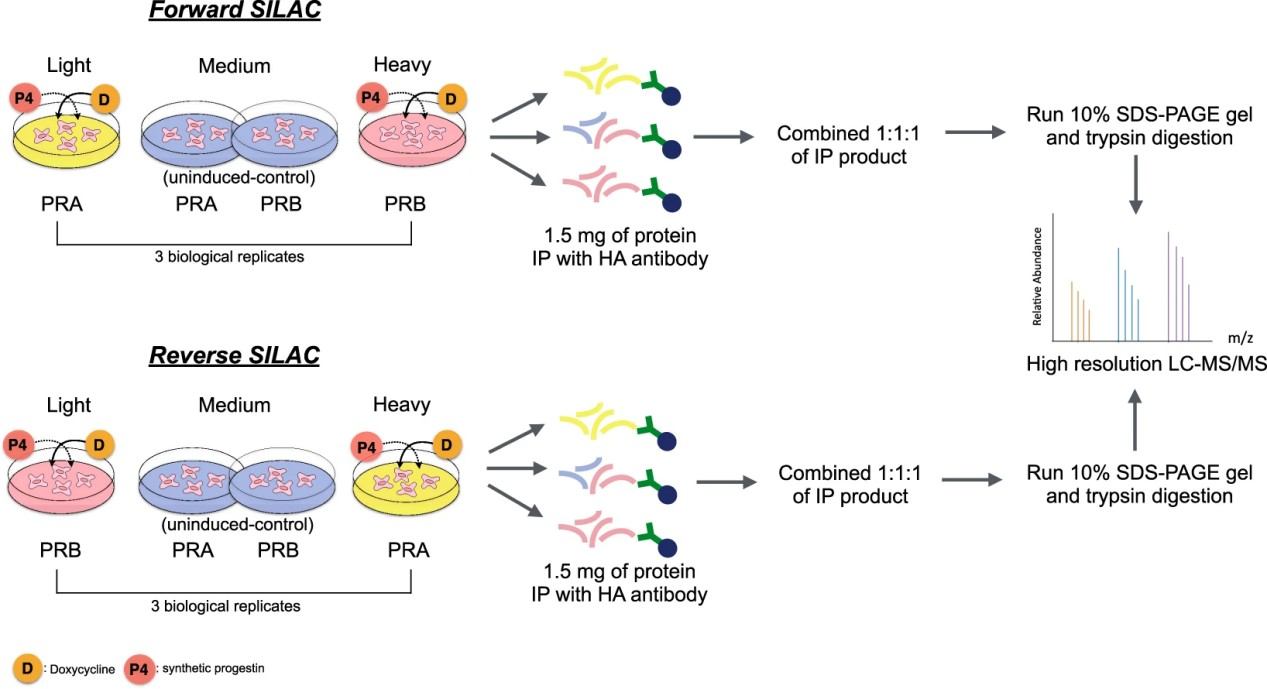
Figure 3. Summary of the experimental workflow applied to investigate PRA and PRB interacting partners.
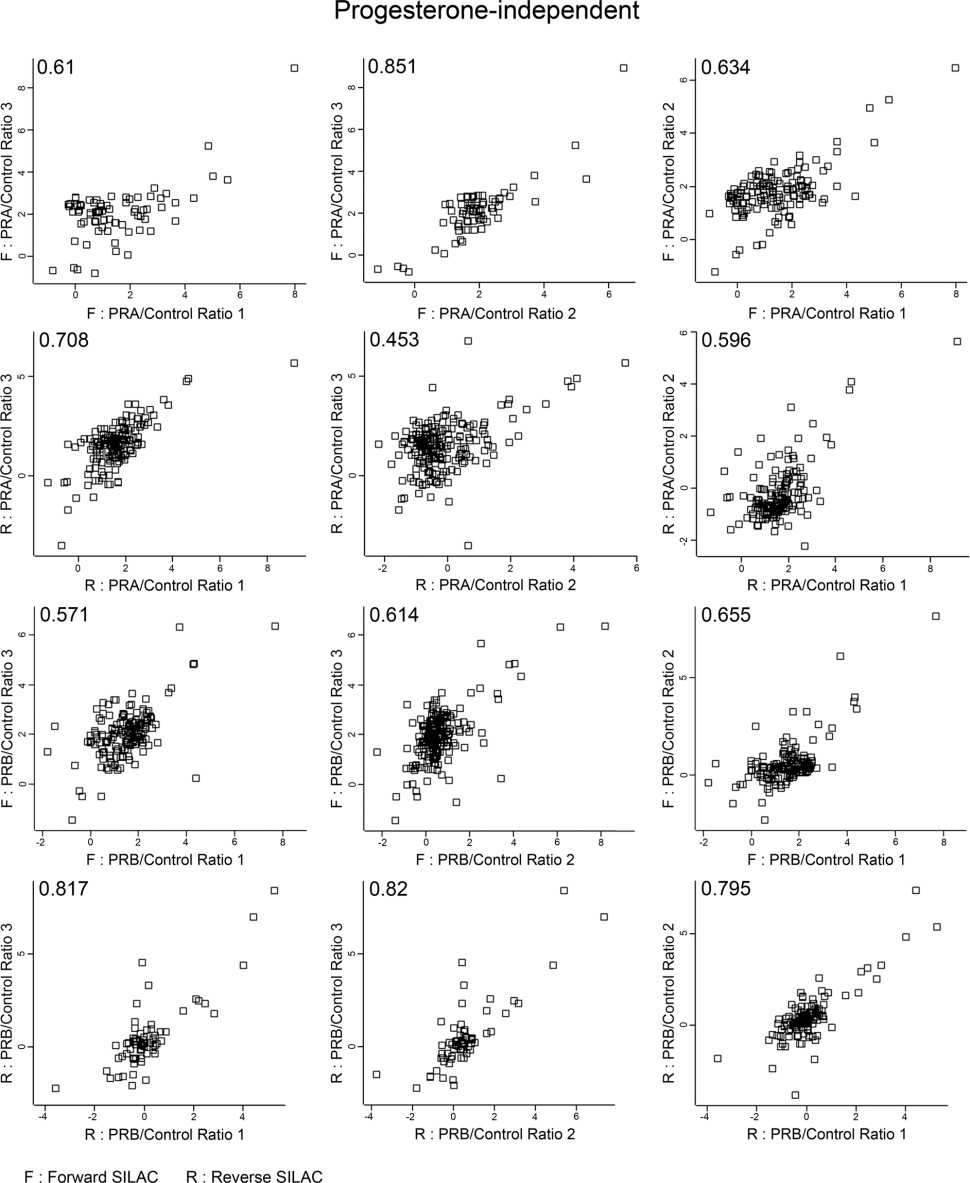
Figure 4. Scatter plots of log2 SILAC ratio comparing across replicates in progesterone-independent condition.
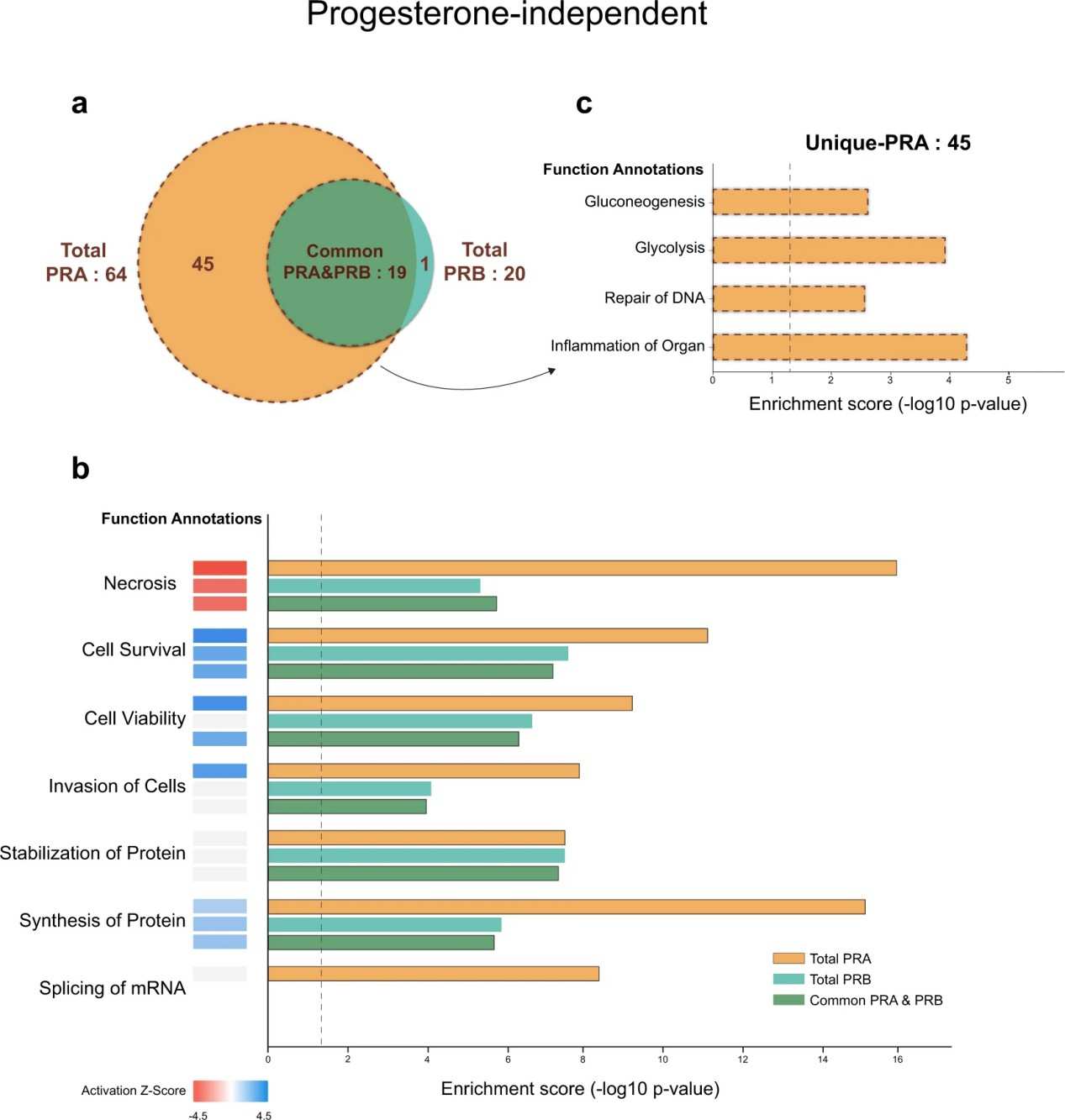
Figure 5. Functional enrichment analysis of progesterone-independent PRA and PRB interacting partners by IPA.
Conclusion
This work demonstrates that SILAC-based Co-IP-MS can resolve subtle quantitative differences in protein-protein interaction networks associated with receptor isoforms and hormonal status. The resulting dataset provides a high-confidence interactome resource for understanding PR-mediated signalling in breast cancer and highlights the value of SILAC for dissecting context-dependent interactomes.
Related Services
References
- Wang X, et al. SILAC–based quantitative MS approach for real-time recording protein-mediated cell-cell interactions. Scientific Reports, 2018, 8(1): 8441.
- Pateetin P, et al. Triple SILAC identified progestin-independent and dependent PRA and PRB interacting partners in breast cancer. Scientific Data, 2021, 8(1): 100.
- Frankenfield A M, et al. Benchmarking SILAC Proteomics Workflows and Data Analysis Platforms. Molecular & Cellular Proteomics, 2025, 24(6).


