Membrane Proteomics
What Is Membrane Proteomics and Why Does It Matter?
Membrane proteomics is a sub-discipline of proteomics dedicated to the large-scale analysis of proteins associated with biological membranes, including plasma membranes and intracellular organellar membranes. Unlike soluble proteins, membrane proteins are embedded within or tightly associated with lipid bilayers, requiring tailored enrichment, solubilization, and analytical strategies.
Understanding the membrane proteome is essential for elucidating cellular signaling networks, transport mechanisms, and protein interaction landscapes. Comprehensive membrane protein profiling enables researchers to investigate how membrane composition varies across different cell types, experimental conditions, or biological states, providing valuable insights into cellular organisation and regulation.
Why Focus on Membrane proteins?
Membrane proteins perform diverse and essential biological functions. They regulate the movement of ions and molecules across membranes, mediate signal transmission between cells and their environment, and facilitate cell–cell recognition and adhesion. These functions position membrane proteins at the interface between intracellular processes and extracellular cues.
From a research perspective, membrane proteins are of particular interest because alterations in their expression, localization, or modification often reflect changes in cellular state. Studying membrane protein dynamics contributes to a deeper understanding of biological systems at both molecular and systems levels, supporting the generation of hypotheses and the exploration of mechanisms.
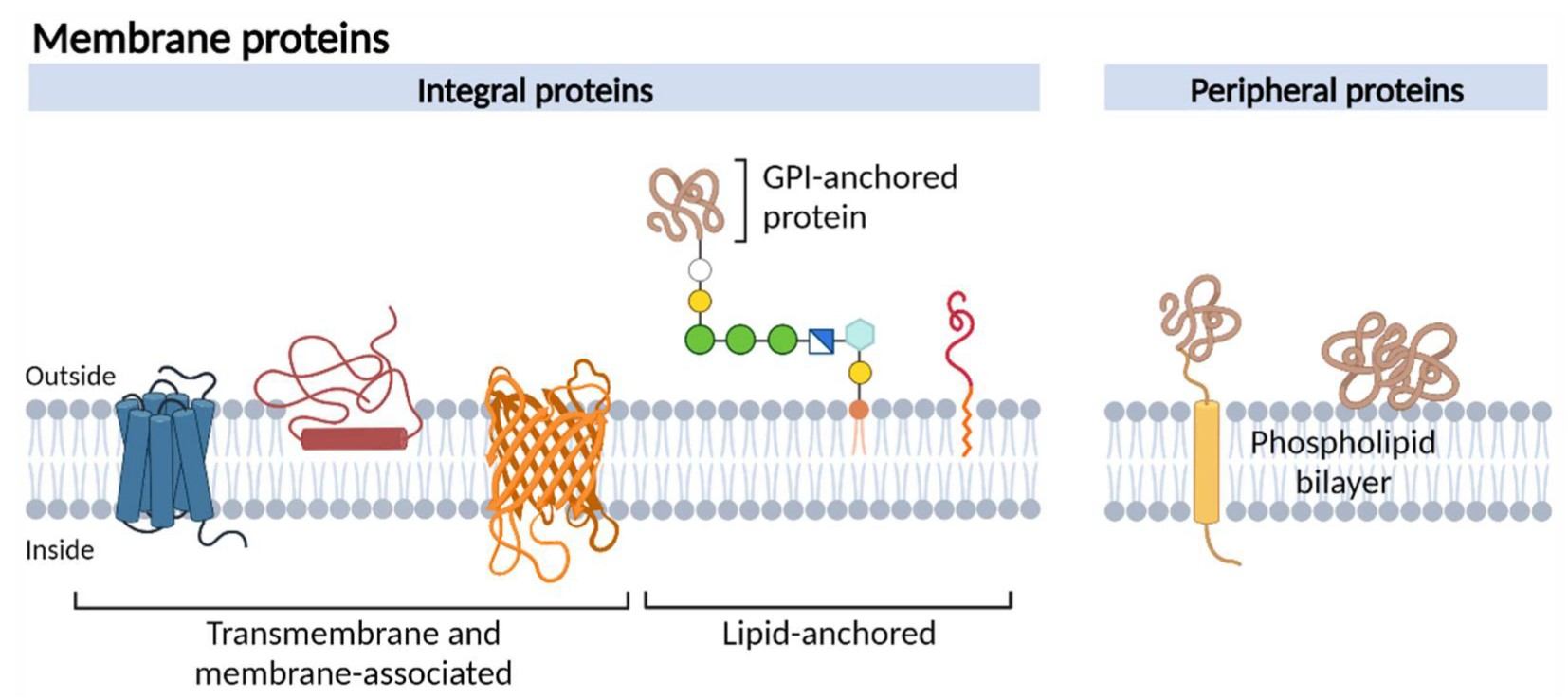
Figure 1. Schematic representation of different membrane protein types (Jong E, Kocer A. 2023).
Membrane enrichment and protein extraction strategies
Effective enrichment reduces dynamic range and improves detection of membrane proteins:
- Density gradient centrifugation: Separates membrane fractions by buoyant density; suitable for plasma membrane and organelle membranes when starting material is abundant.
- Affinity-based enrichment: Antibody-capture using immobilized antibodies or lectin affinity for subpopulations of surface proteins.
- Surface labelling and capture: Chemical labelling of extracellular domains followed by selective capture enriches cell surface proteins while permitting stringent washing to remove associated cytosolic contaminants.
For extraction and solubilisation:
- Use detergent or organic solvent systems validated to solubilise transmembrane domains while remaining compatible with downstream LC–MS/MS.
- Consider sequential extraction (mild to strong) to fractionate peripheral vs integral membrane proteins.
- When intact-protein analysis is planned, adjust solubilisation to preserve native mass and PTMs.
Protein identification by advanced LC–MS/MS
High-resolution LC–MS/MS is the workhorse for the identification of membrane proteins. Key considerations:
- Peptide-centric (bottom-up) workflows provide broad coverage; hydrophobic peptides often require optimized chromatographic gradients and pre-fractionation.
- Intact-protein (top-down) approaches can resolve proteoforms and post-translational modifications directly, valuable when PTM mapping is a primary objective.
- Instrument platform features such as high resolving power, accurate mass, and sensitive fragmentation methods improve identification confidence for low-abundance membrane proteins.
Quantitative Membrane Proteomics: Strategy Selection
Quantitative analysis is crucial for comparing the abundance of membrane proteins across samples or experimental conditions. Creative Proteomics' Membrane Proteomics services provide flexible quantitative strategies to accommodate various study designs and throughput requirements.
Label-Based Quantification Approaches
Label-based strategies enable accurate relative quantification by incorporating stable isotopes or chemical tags during sample preparation. Use chemical or metabolic labels when you need precise, relative comparisons across a small set of samples (for example, 4–16 conditions). Labels let you run samples together, reducing run-to-run variation and improving fold-change accuracy. Best for discovery experiments where precise control of technical variability is crucial.
Label-Free Quantification Approaches
Label-free strategies rely on signal intensity or targeted monitoring to quantify proteins across samples without chemical labeling. Techniques such as targeted and data-independent acquisition support scalable analysis and are well suited for verification studies or large sample sets. Label-free methods provide flexibility while maintaining quantitative robustness.
- Targeted methods (SRM/MRM, PRM): Allow sensitive, reproducible quantification of preselected peptides; ideal for verification of biomarker candidates.
- Data-independent acquisition (DIA): Provides reproducible, broad coverage across cohorts without labels; suitable for larger-scale profiling where consistent sampling depth is required.
Deliverables and Reporting Standards
- Detailed experimental protocol and QC metrics.
- Raw and processed LC–MS/MS data and identification/quantification tables.
- Annotated lists of membrane proteins with topology and functional annotation.
- Publication-ready visualisations (heatmaps, volcano plots, interaction networks) for research dissemination.
Bioinformatics, Topology Prediction, and Functional Interpretation
Bioinformatics turns identifications into biological insight. Essential analysis components include:
- Subcellular localisation and transmembrane topology prediction to confirm membrane assignment and infer orientation.
- Gene Ontology (GO) and pathway (e.g., KEGG) enrichment to identify over-represented biological processes.
- PPI networks to prioritise proteins with central roles or potential regulatory connections.
- Proteoform and PTM mapping to detect modification-dependent regulation.
Post-Translational Modification (PTM) Analysis of Membrane Proteins
Membrane proteomics workflows can be adapted to systematically detect and characterize PTMs, including phosphorylation, glycosylation, ubiquitination, and lipidation, which regulate membrane protein localization, stability, and signaling interactions. From a methods perspective, PTM analysis typically begins with optimized membrane enrichment and extraction protocols that preserve labile modifications.
- Phosphorylation: Phosphopeptide enrichment techniques combined with high-resolution LC–MS/MS allow site-specific mapping of low-abundance phosphorylated residues on receptors and transporters.
- Glycosylation: Glycosylation analysis often involves glycopeptide capture, deglycosylation-assisted site mapping, and intact glycopeptide profiling to define site occupancy and glycan heterogeneity.
- Lipid: Lipid modifications, such as acylation or prenylation, are captured using hydrophobic-compatible extraction and MS-compatible enrichment to preserve membrane association.
Creative Proteomics' Membrane Proteins Service Workflow
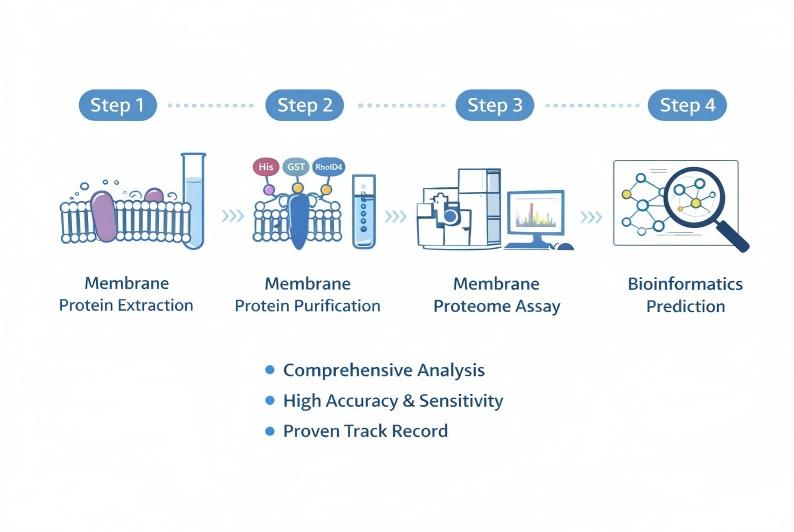
Representative Applications
Membrane proteomics supports a range of laboratory research objectives, for example:
- Receptor and transporter profiling to elucidate changes in signalling landscapes under experimental perturbations.
- Biomarker candidate discovery in model systems or preclinical samples.
- Mechanistic studies of membrane-associated pathways using integrated proteomics and functional assays.
- Resistance-mechanism research by comparing membrane proteomes between perturbed and control models.
Sample Requirements for Membrane Proteomics Analysis
| Sample Type | Recommended Amount | Minimum Amount | Preparation & Storage Notes |
| Cultured cells | ≥ 1 × 10⁷ cells | 5 × 10⁶ cells | Wash with cold PBS, pellet cells, snap-freeze in liquid nitrogen; store at −80 °C. |
| Animal or plant tissue | 50–200 mg | 20 mg | Remove excess fat/connective tissue; snap-freeze immediately; store at −80 °C. |
| Plasma membrane fraction | ≥ 50 µg protein | 20 µg protein | Isolate using validated enrichment protocols; avoid repeated freeze–thaw cycles. |
| Exosomes / extracellular vesicles | ≥ 1 × 10¹⁰ particles | 5 × 10⁹ particles | Purify vesicles prior to submission; resuspend in PBS; store at −80 °C. |
| Bacterial cells | ≥ 1 × 10⁹ cells | 5 × 10⁸ cells | Pellet cells, wash with PBS, snap-freeze; store at −80 °C. |
| FFPE tissue sections | ~80 mg tissue (≥4 sections, 5–20 µm) | 40 mg tissue | Ensure proper fixation; ship at room temperature if fully fixed. |
Why Choose Creative Proteomics for Membrane Proteomics Partner?
- Proven expertise with hydrophobic and low-abundance proteins
- Access to high-resolution MS platforms
- Transparent QC and bioinformatics pipelines
- Flexible, project-specific workflow design
- Experience supporting pharma and CRO timelines
FAQ
-
Q1: What types of membrane proteins can be reliably identified using membrane proteomics?
A1: Membrane proteomics enables the identification of integral transmembrane proteins, peripheral membrane proteins, lipid-anchored proteins, and membrane-associated signaling complexes, including GPCRs, ion channels, transporters, receptors, and adhesion molecules.
-
Q2: How does membrane protein extraction for proteomics differ from standard protein extraction?
A2: Extraction methods for membrane proteomics often incorporate detergents or organic solvents that maintain membrane protein solubility while minimizing interference with downstream analyses. Specialised sample preparation techniques (e.g., filter-aided sample preparation, SP3) can help mitigate detergent contamination and enhance mass spectrometry compatibility.
-
Q3: How does membrane proteomics integrate with broader systems biology?
A3: Membrane proteomics data are often combined with transcriptomics, phosphoproteomics, and functional assays to build comprehensive models of cellular regulation, signaling cascades, and drug response networks. Coupled with bioinformatics annotation, this integrative approach enhances mechanistic understanding.
-
Q4: How does membrane proteomics contribute to drug resistance studies?
A4: By comparing membrane protein expression and modification profiles between drug-sensitive and drug-resistant models, membrane proteomics can reveal altered transporters, receptors, or signaling components that underlie therapeutic resistance mechanisms.
Demo
Demo: Analysis of differential membrane proteins related to matrix stiffness-mediated metformin resistance in hepatocellular carcinoma cells
This study employed iTRAQ quantitative membrane proteomics to investigate the impact of extracellular matrix stiffness on the expression of membrane proteins and metformin resistance in liver cancer cells. The analysis revealed specific, differentially regulated membrane proteins associated with drug response mechanisms, highlighting the utility of membrane proteomics in studying therapeutic resistance.
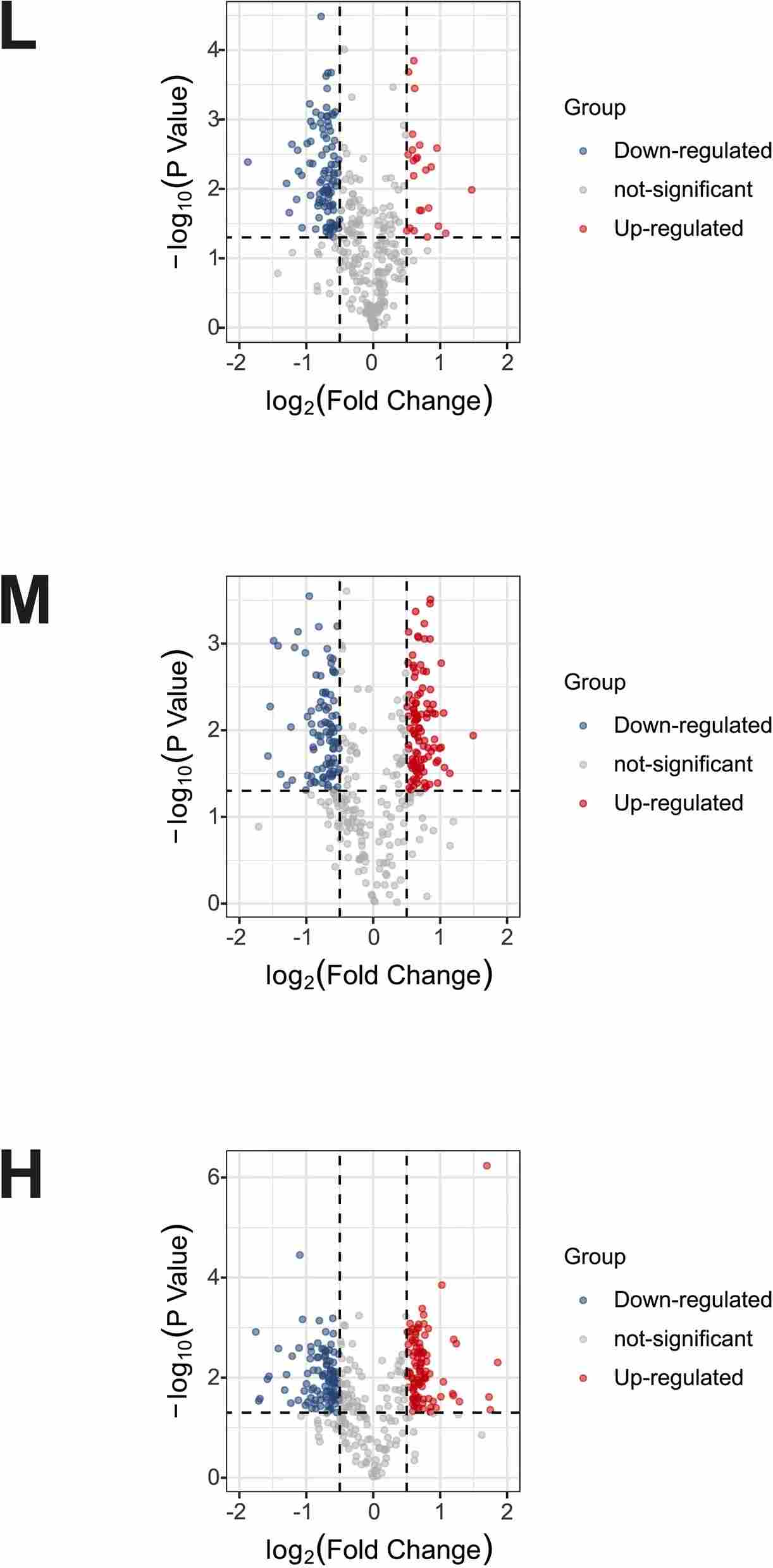
Figure 2. The volcano plot of differentially expressed membrane proteins.
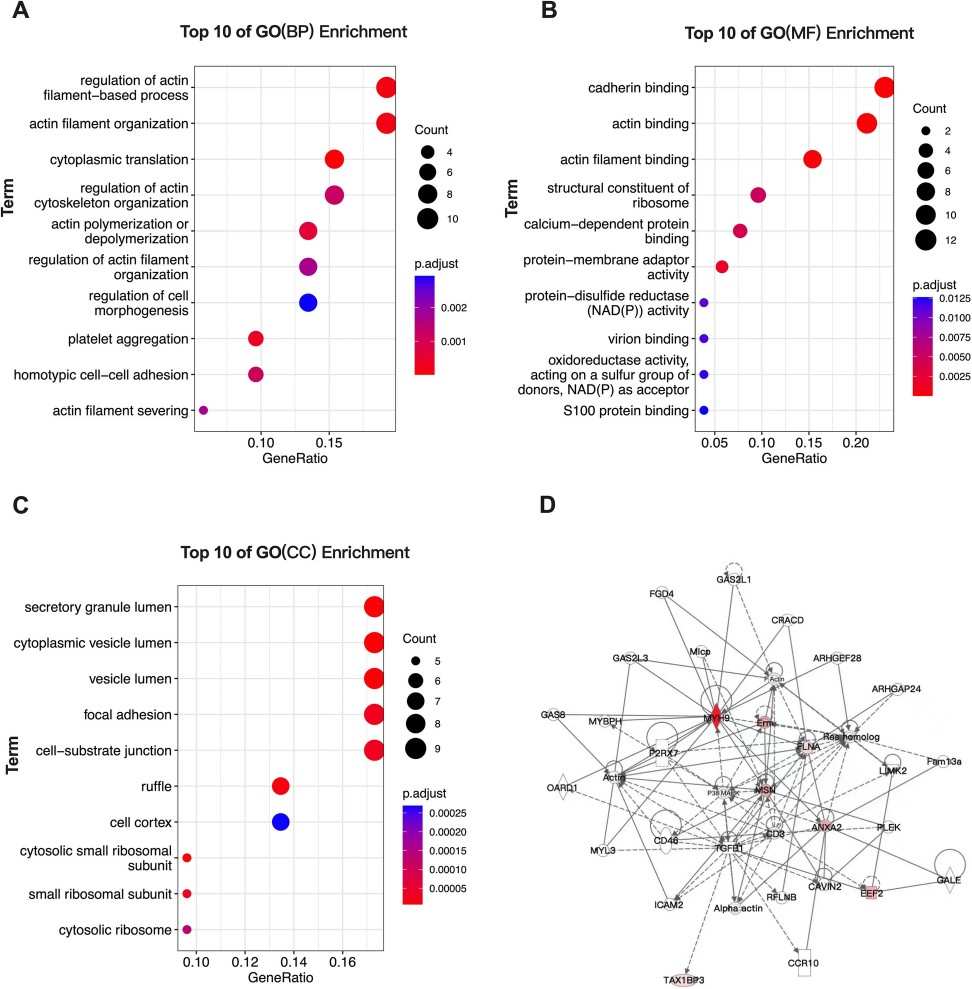
Figure 3. Gene Ontology (GO) enrichment analysis of genes in cluster 1 and construction of a PPI network for the six candidate membrane proteins.
-
Case Study
Case: Cell Surface Proteomics Reveals Hypoxia‑Regulated Pathways in Cervical and Bladder Cancer.
Background
Plasma membrane proteins are critical for cell signalling, adhesion, and trafficking, and because they reside at the cell surface, they are prime therapeutic targets in cancer research. However, their hydrophobicity and low abundance make them difficult to detect with conventional proteomic techniques.
Purpose
This study aimed to enhance the detection and identification of hypoxia-responsive membrane proteins in cervical (SiHa) and bladder (UMUC3) cancer cell lines by combining cell surface biotinylation enrichment with LC-MS/MS proteomics, thereby enabling the discovery of proteins and pathways not captured by standard whole-cell analysis.
Methods
- Cell models: Human cervical (SiHa) and bladder (UMUC3) cancer cells were cultured under normoxic and hypoxic conditions to mimic tumor microenvironments.
- Membrane enrichment: Cell surface proteins were selectively labelled with a biotinylation reagent that only reacts with extracellular protein residues, followed by affinity purification to isolate enriched plasma membrane proteins.
- Proteomic analysis: Enriched membrane proteins were digested and identified using LC–MS/MS.
- Data analysis: Differential protein abundance and pathway enrichment were determined by comparing hypoxia vs. normoxia conditions across biotin‑enriched membrane fractions and whole‑cell lysates.
Results
- Hypoxia‑specific membrane proteins: In SiHa cells, 43 hypoxia‑upregulated proteins were identified exclusively in the biotin‑enriched fraction, including integrins and receptor kinases.
- In UMUC3 bladder cancer cells, 32 unique hypoxia‑responsive membrane proteins were detected only after biotin enrichment.
- Pathway insights: Identified proteins were linked to extracellular matrix remodelling, integrin signalling, PI3K–Akt signalling, and immune modulation.
- Protein interactions: Network analysis revealed connections between membrane receptors and intracellular stress regulators, suggesting coordinated regulation under hypoxia.
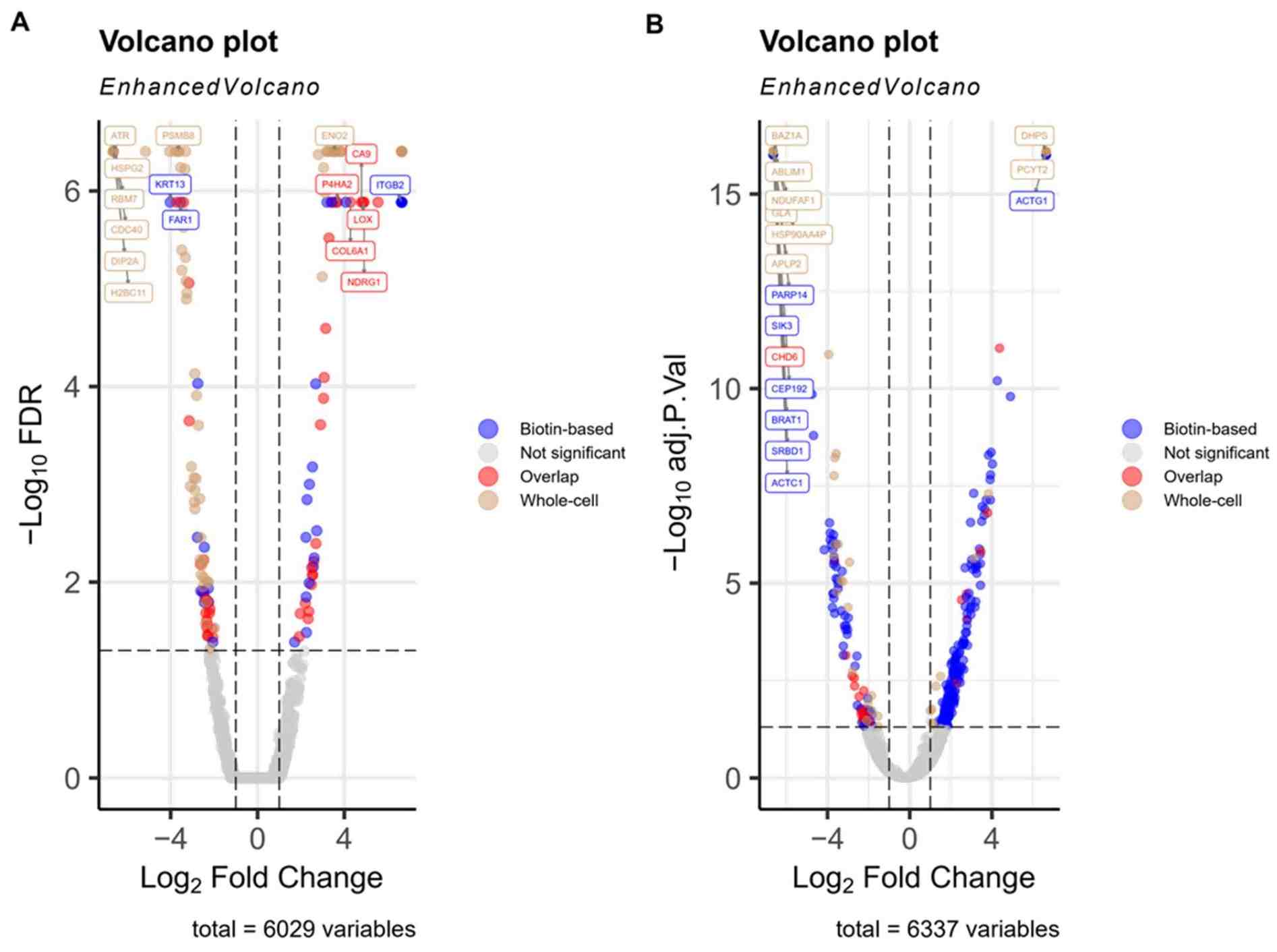
Figure 4. Volcano plots of differentially abundant proteins under hypoxia in SiHa and UMUC3 cells.
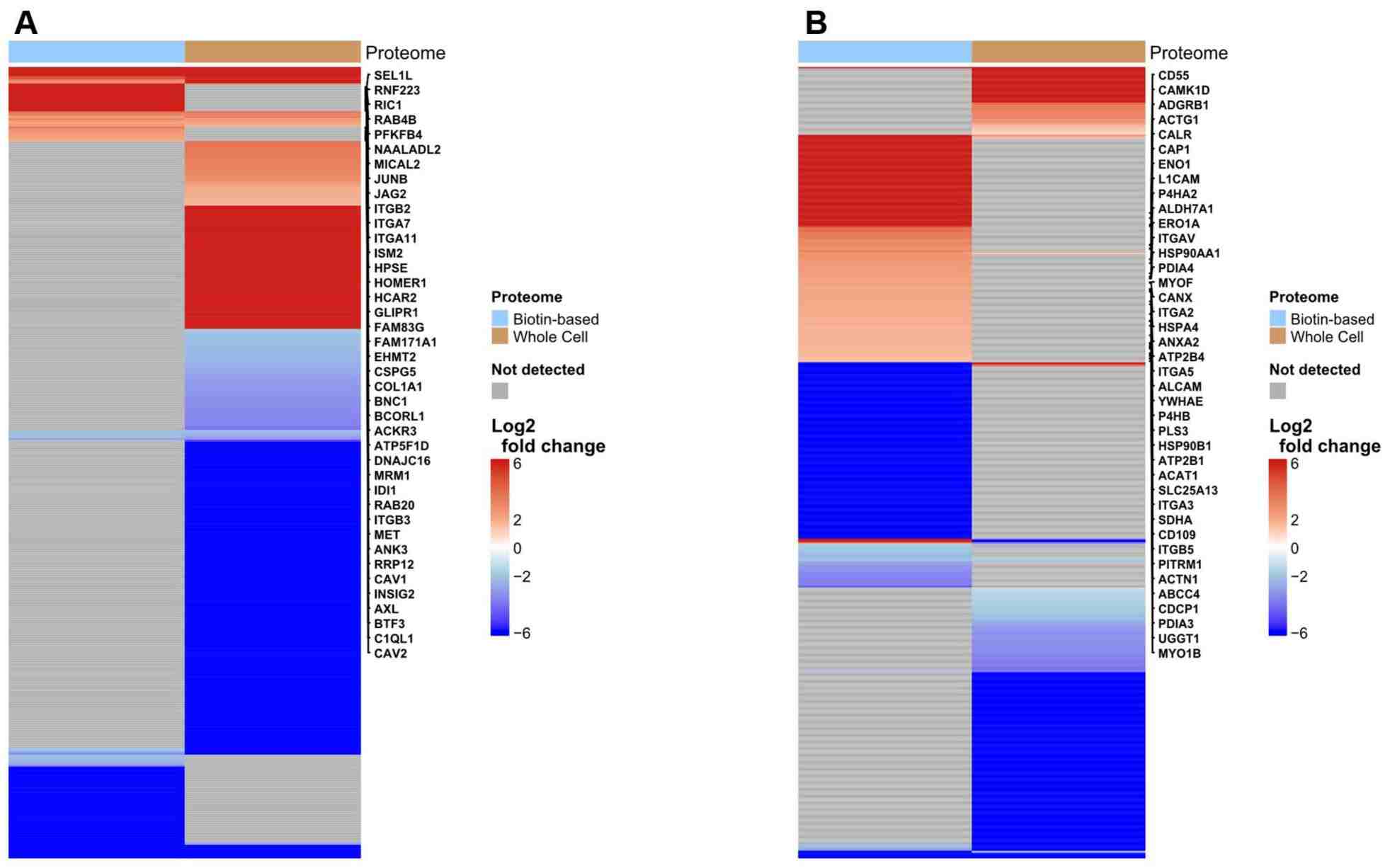
Figure 5. Heatmaps of differentially abundant proteins in biotin-enriched and whole-cell fractions of SiHa and UMUC3 cells under hypoxia.
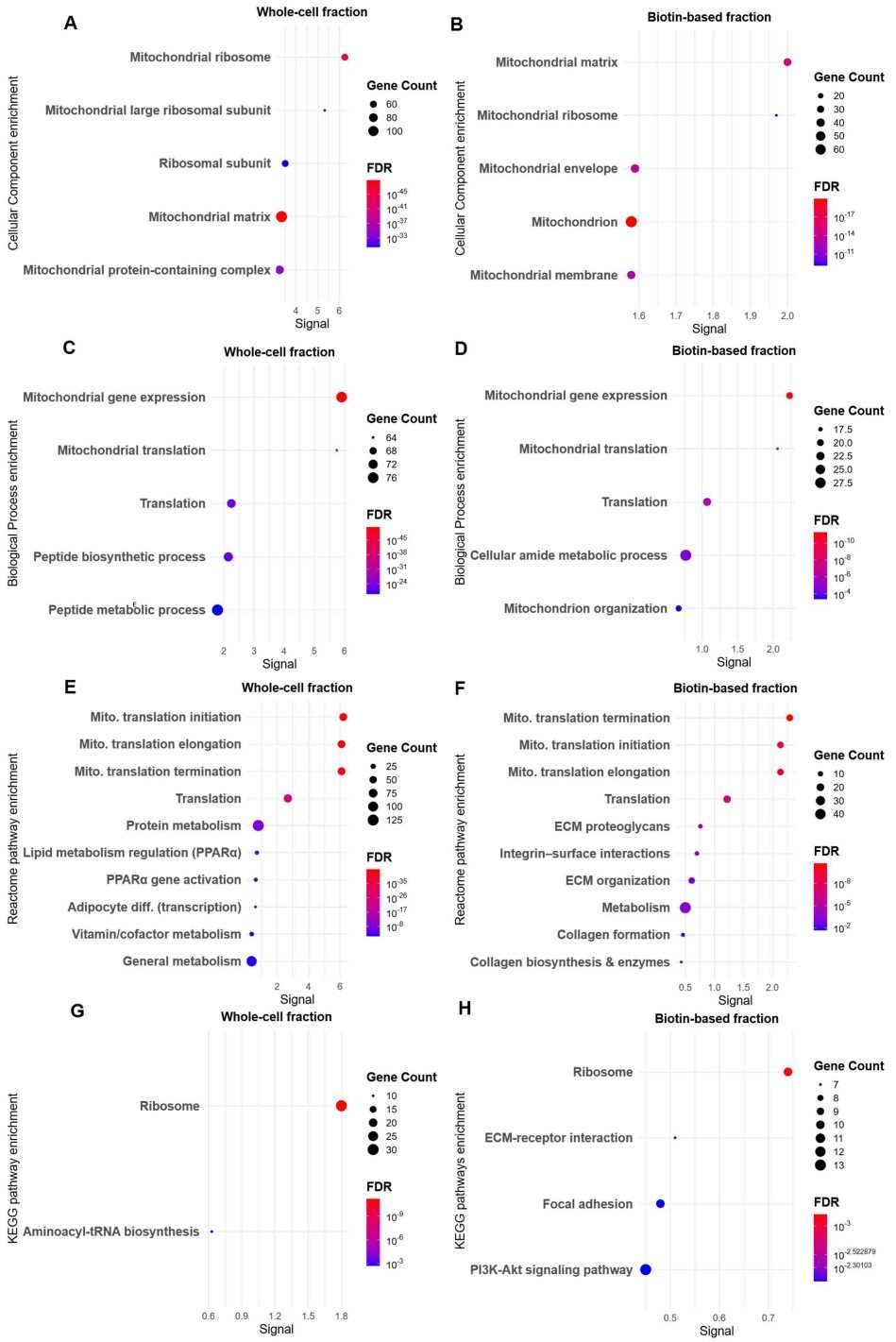
Figure 6. GO and pathways enrichment analyses of differentially abundant proteins in SiHa cervical cancer cells under hypoxia.
Conclusion
The incorporation of cell surface biotinylation enrichment before LC-MS/MS dramatically improves the sensitivity and specificity of plasma membrane proteome profiling in cancer models, particularly under stress conditions such as hypoxia. This strategy revealed hypoxia‑regulated membrane proteins and signalling pathways that traditional whole‑cell proteomics missed, highlighting the value of targeted membrane enrichment in identifying novel therapeutic targets and biomarkers.
Related Services
References
- Jong E, Kocer A. Current methods for identifying plasma membrane proteins as cancer biomarkers. Membranes, 2023, 13(4): 409.
- Gao X, et al. Analysis of differential membrane proteins related to matrix stiffness-mediated metformin resistance in hepatocellular carcinoma cells. Proteome Science, 2023, 21(1): 14.
- Alanazi F, et al. Cell Surface Proteomics Reveals Hypoxia-Regulated Pathways in Cervical and Bladder Cancer. Proteomes, 2025, 13(3): 36.


