Chemical Proteomics Service
What Is Chemical Proteomics?
Chemical proteomics is an advanced branch of proteomics that systematically investigates interactions between small molecules and proteins at a proteome-wide scale. By integrating chemical biology strategies with high-resolution mass spectrometry and quantitative data analysis, chemical proteomics enables unbiased identification of molecular targets, binding partners, and affected pathways within complex biological systems.
Unlike traditional hypothesis-driven approaches, chemical proteomics does not rely on prior assumptions about targets. Instead, it offers a comprehensive and experimentally grounded perspective on how small molecules interact with the proteome in a biologically relevant context. It makes chemical proteomics a powerful research tool for target discovery, elucidation of the mechanism of action, and characterization of compounds across early-stage biomedical research and pharmaceutical development workflows.
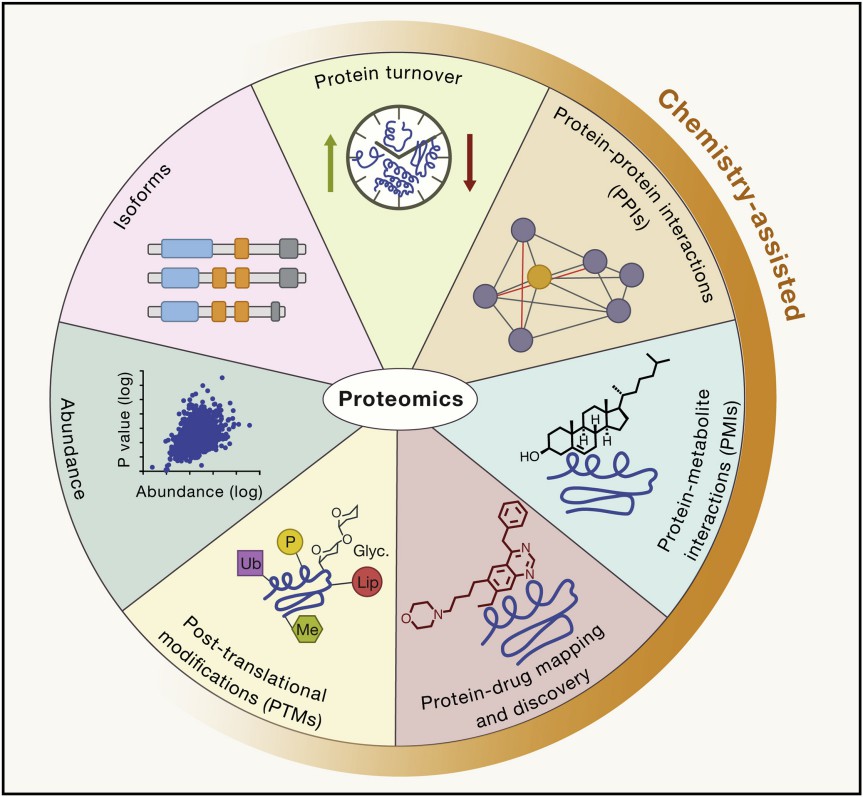
Figure 1. Applications that can be integrated with chemical methods to aid in proteomic investigations (Bantscheff M, et al., 2012).
Why Chemical Proteomics Matters in Drug Discovery Research?
Understanding how small molecules interact with biological systems remains one of the most critical challenges in modern drug discovery research. Many compounds exhibit complex biological effects that cannot be explained by single-target models alone. Chemical proteomics addresses this challenge by providing an unbiased strategy to identify both primary and secondary protein interactions under near-physiological conditions. By enabling systematic target deconvolution, chemical proteomics helps:
- Identify unknown or unexpected protein targets
- Assess compound selectivity and off-target engagement
- Explore downstream pathway perturbations
- Support structure–activity relationship (SAR) analysis
Principles of Chemical Proteomics Analysis
Proteome-Wide Small Molecule Interaction Mapping
At the core of chemical proteomics is the ability to capture and identify proteins that directly or indirectly interact with a given compound. Depending on experimental design, this interaction may be detected through physical binding, changes in protein stability, altered abundance, or shifts in post-translational modification states.
Quantitative Mass Spectrometry
High-resolution LC-MS/MS serves as the analytical foundation of chemical proteomics. Quantitative MS strategies enable precise comparison between treated and control samples, allowing for the statistically robust identification of compound-responsive proteins.
Chemical Proteomics Strategies We Support
| Chemical Proteomics Strategy | Principle | Application | Advantages |
| Affinity-Based Chemical Proteomics | The compound is attached to a solid support and used as bait to "fish out" proteins that bind to it. | When the compound can be modified without losing activity and direct binding partners are needed. | High confidence target identification.
Enables discovery of protein complexes. |
| Label-Free Chemical Proteomics | Protein behavior changes after compound treatment reveal likely interactions. | When chemical modification is not feasible or native compound structure must be preserved. | Maintains biological relevance.
Suitable for natural products and early discovery. |
| Thermal and Stability-Based Proteomics (e.g., TPP, PISA) | Protein binding alters stability, detected by changes under heat or chemical stress. | When unbiased, proteome-wide target discovery is required without probe design. | No probe needed.
Supports multiplexed and quantitative analysis. |
| Covalent and Reactivity-Based Chemical Proteomics | Reactive compounds form stable bonds with specific protein sites. | When studying electrophilic or covalent compounds and selectivity is critical. | Identifies binding sites.
Reveals off-target reactivity and engagement patterns. |
Integration with Other Proteomics and Omics Platforms
Chemical Proteomics and PTM Profiling
Combining chemical proteomics with PTM profiling connects who a small molecule touches with what it changes inside the cell. Chemical proteomics identifies the proteins that bind to a compound; PTM profiling maps changes in chemical tags added to proteins (for example, phosphate groups, small protein tags that mark proteins for removal, or acetyl groups that alter activity). Together, they distinguish direct binding events from downstream signaling responses and reveal how a compound rewires pathways.
Multi-Omics Data Integration
By integrating proteomics with transcriptomics, metabolomics, and epigenomics, researchers can see how a small molecule affects entire biological systems rather than just individual proteins. For example, changes in protein abundance can be cross-referenced with gene expression data to determine whether effects are driven at the transcriptional level or post-translationally. Similarly, metabolomics data can reveal how altered proteins influence metabolic pathways, highlighting downstream effects on cell energy, signaling, or stress responses.
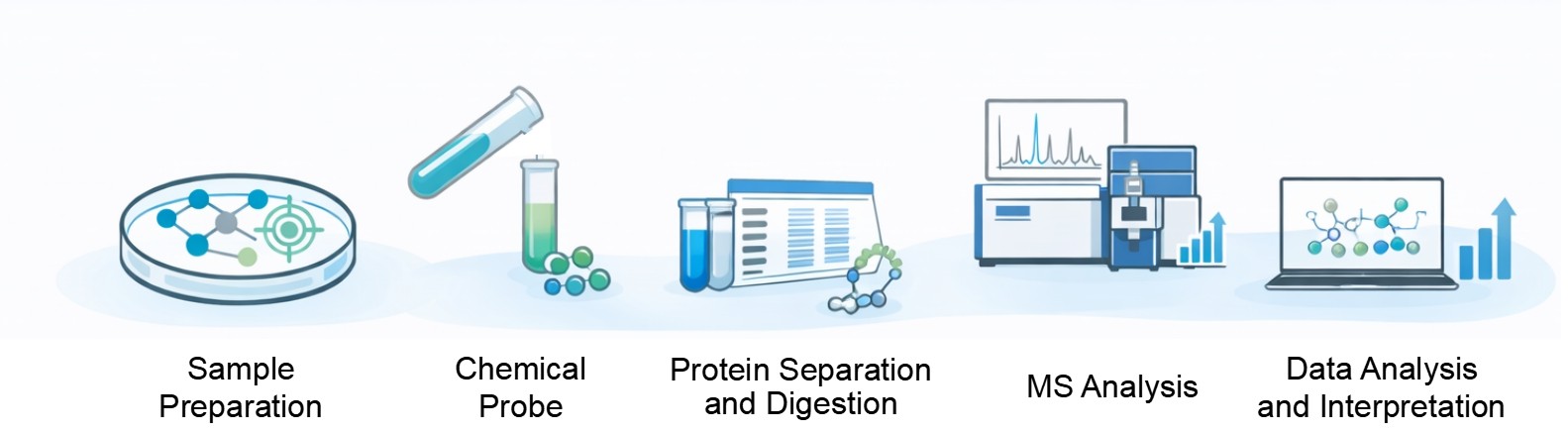
Our Chemical Proteomics Service Workflow
- Sample Preparation: Cells, tissues, or biological fluids are carefully processed to preserve protein integrity and functional states.
- Chemical Probe Application: Depending on your project, introducing small molecules or probes to the prepared samples.
- Protein Separation and Digestion: Proteins are separated to reduce sample complexity, using techniques such as gel electrophoresis or liquid chromatography. Separated proteins are then digested into smaller peptides.
- Mass Spectrometry Analysis: Quantitative methods reveal differences in protein abundance, stability, or interaction patterns between treated and control samples.
- Data Analysis and Interpretation: Specialized software and statistical models process the mass spectrometry data to identify proteins that interact with the compound, respond to treatment, or show altered stability.
Data Analysis and Bioinformatics for Chemical Proteomics
- Quantitative data processing and quality control: Raw mass spectrometry files are processed to identify and quantify proteins across all experimental conditions.
- Statistical identification of compound-responsive proteins: Proteins are compared between treated and control samples.
- Target prioritization and ranking: Candidate targets are ranked based on multiple criteria.
- Pathway and network-level interpretation: Identified proteins are mapped to biological pathways and interaction networks.
- Integration with complementary proteomics data: Chemical proteomics results can be integrated with quantitative expression data, protein modification profiles, or stability-based measurements.
Applications of Chemical Proteomics Services
Target Identification and Deconvolution
Chemical proteomics can reveal which proteins a small molecule directly binds to in cells or tissues. It enables researchers to identify both known and unexpected targets, thereby clarifying why a compound produces specific cellular effects.
Mechanism-of-Action and Pathway Mapping
Beyond identifying targets, chemical proteomics shows how a molecule affects cellular pathways. Changes in protein abundance, stability, or interaction networks can reveal which biological processes are influenced, such as cell signaling, metabolism, or stress responses.
Selectivity and Off-Target Profiling
Chemical proteomics can detect unintended interactions, highlighting proteins that may contribute to side effects or reduced efficacy. This helps researchers refine compound design and improve selectivity before moving to more complex studies.
Compound Ranking and Lead Optimization
By comparing protein engagement profiles across related compounds, chemical proteomics guides the prioritization of molecules with the most promising activity and minimal off-target effects.
Support for Functional and Phenotypic Studies
Chemical proteomics data complement other cellular or functional assays. For example, linking protein target profiles to observed changes in cell growth, signaling, or stress responses can validate hypotheses about compound activity, providing a stronger foundation for mechanistic studies.
Sample Requirements for Chemical Proteomics Services
| Sample Type | Recommended Quantity / Volume | Preparation Notes |
| Cultured Cells | 1-50 million cells | Cells should be harvested under defined culture conditions; wash to remove serum or media components; lyse promptly to preserve native proteome |
| Primary Cells | 0.5-10 million cells | Minimize time from isolation to lysis; gentle handling to preserve functional protein complexes |
| Tissue Samples | 10-100 mg | Snap-freeze tissue immediately after collection; avoid repeated freeze–thaw; homogenize in protease inhibitor–containing lysis buffer |
| Biofluids (Plasma, Serum, Urine) | 100 µL-5 mL | Clarify samples by centrifugation; remove abundant proteins if necessary for low-abundance target detection |
Why Choose Creative Proteomics for Chemical Proteomics Services
- End-to-End Experimental Design: We design studies around your specific scientific question, carefully selecting probe strategies, sample types, and controls to minimize artifacts and maximize meaningful results.
- High-Resolution Proteome Analysis: Using state-of-the-art mass spectrometry and multiplexed quantification, we can measure thousands of proteins with high accuracy and reproducibility, capturing both global abundance and post-translational modifications.
- Actionable Biological Insights: Our team generates clear, interpretable outputs, including ranked target lists with confidence scores, pathway maps illustrating downstream effects, and detailed information on protein binding sites or reactive residues.
- Comprehensive Data Delivery: All projects include processed results, raw files, and detailed analysis notes. Flexible engagement models allow reproducibility, reanalysis, and seamless integration into ongoing research workflows.
FAQ
-
Q1: Why is probe design critical in chemical proteomics experiments?
A1: Probes must retain the small molecule's biological activity while incorporating reactive groups and reporter tags that allow efficient target capture and enrichment.
-
Q2: Can chemical proteomics identify both on-target and off-target interactions?
A2: Yes. Chemical proteomics can reveal both primary targets and secondary or off-target interactions by profiling interaction signatures across the proteome.
-
Q3: How do thermal profiling methods contribute to chemical proteomics?
A3: Thermal methods detect shifts in protein thermal stability upon ligand binding, enabling target identification without probe modification.
Demo
Demo: Target identification of natural medicine with chemical proteomics approach: probe synthesis, target fishing and protein identification
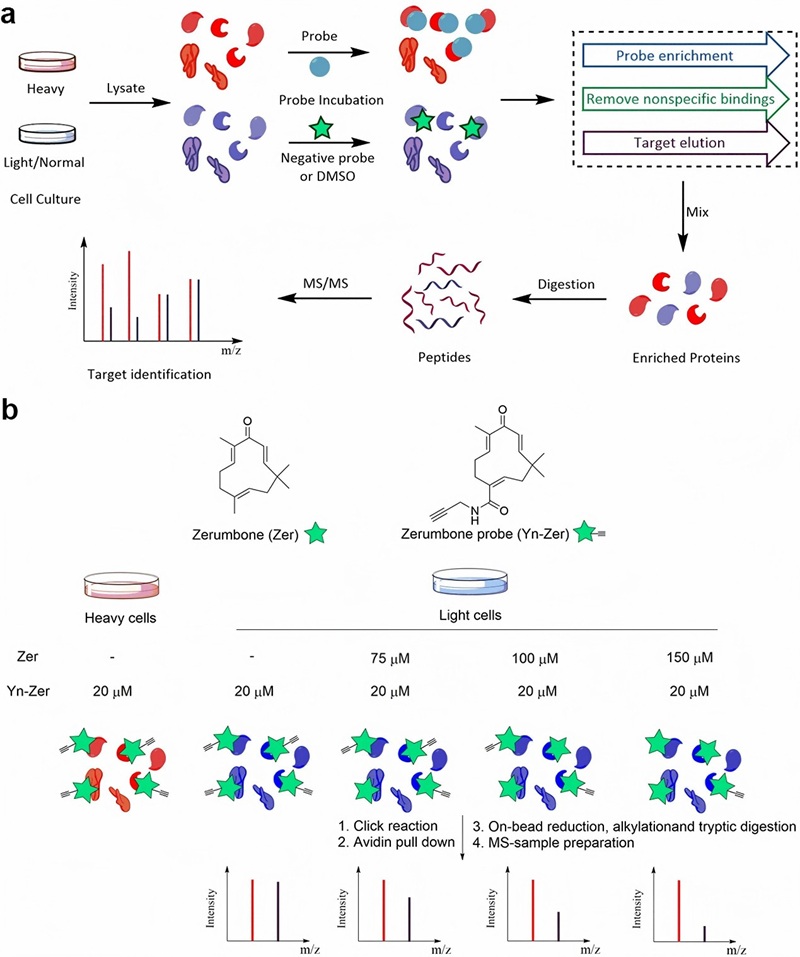
Figure 2. Target identification of zerumbone through a chemical proteomics approach coupled with SILAC (Chen X, et al., 2020).
-
Case Study
Case: Chemical Proteomics Identifies Nampt as the Target of CB30865, An Orphan Cytotoxic Compound
Author: Chen X, et al.
Journal: Signal Transduction and Targeted Therapy
Publication: 2020
DOI: 10.1038/s41392-020-0186-y
Background
Phenotype-driven drug discovery frequently identifies bioactive small molecules whose molecular targets remain unknown, limiting further optimization and mechanistic understanding. CB30865 is a highly potent cytotoxic compound discovered in the late 1990s. Despite extensive pharmacological, genomic, and resistance profiling efforts, its mechanism of action and direct cellular target remained unresolved for over a decade.
Purpose
Identify the molecular target of CB30865 and determine whether its cytotoxicity can be mechanistically explained by direct inhibition of a specific protein; validate the target biochemically and in cells, and evaluate whether target engagement explains the SAR and cellular phenotypes.
Methods
- Performed small-molecule affinity purification: immobilized MPI-0479883 on beads, incubated with cell lysates, separated by SDS-PAGE, and identified enriched proteins by nanoLC–MS/MS followed by spectral counting and hierarchical clustering. Confirmed hits by immunoblotting.
- Assessed biochemical inhibition using a coupled in-vitro Nampt assay; used a counter-assay including NMN to bypass Nampt.
- Performed cellular mechanism assays: (1) PAR immunofluorescence after H₂O₂ to monitor PARP activity, (2) direct measurement of cellular NAD after 24h treatment, and (3) ATP-based viability with and without nicotinic acid (NA) rescue.
- Compared potencies across compounds and controls and correlated pull-down specificity with cellular potency and SAR.
Results
- Chemical proteomics revealed nicotinamide phosphoribosyltransferase (Nampt) as the sole high-confidence binding protein of CB30865 derivatives.
- CB30865 analogs inhibited Nampt activity at subnanomolar concentrations, leading to depletion of intracellular NAD levels, suppression of PAR formation following DNA damage, and loss of cell viability.
- All cellular effects were fully reversed by supplementation with nicotinic acid, demonstrating that cytotoxicity was directly attributable to Nampt inhibition.
- Structure-activity relationships across multiple analogs consistently correlated Nampt inhibition with cytotoxic potency.
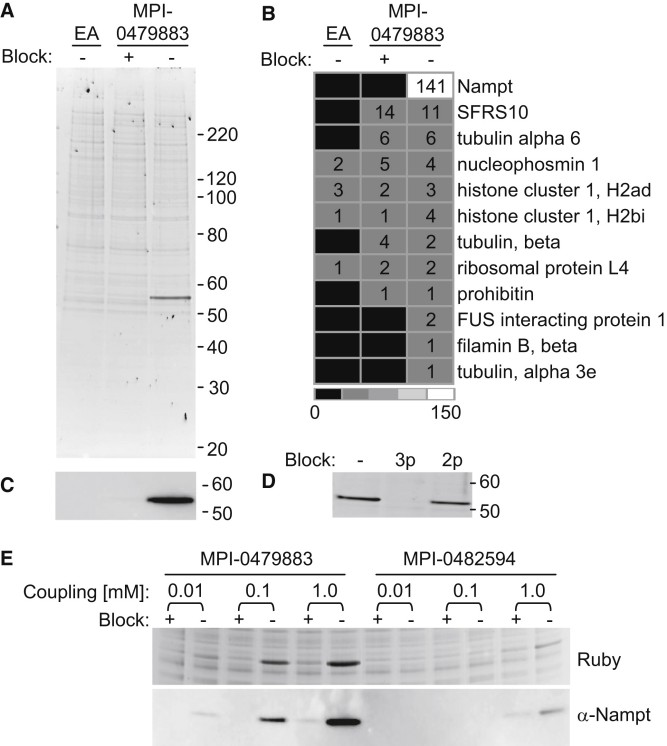
Figure 3. MPI-0479883 Interacts with Nampt.
Conclusion
This study conclusively identified Nampt as the molecular target of CB30865, thereby resolving a long-standing mystery surrounding the orphan compound mechanism using chemical proteomics. The findings validate chemical proteomics as a robust and unbiased approach for deconvoluting the targets of small molecules with unknown modes of action. Additionally, the unique chemical scaffold of CB30865 provides new starting points for future exploration of Nampt biology and the development of inhibitors.
Related Services
References
- Parker C G, Pratt M R. Click chemistry in proteomic investigations. Cell, 2020, 180(4): 605-632.
- Chen X, et al. Target identification of natural medicine with chemical proteomics approach: probe synthesis, target fishing and protein identification. Signal Transduction and Targeted Therapy, 2020, 5(1): 72.
- Fleischer T C, et al. Chemical proteomics identifies Nampt as the target of CB30865, an orphan cytotoxic compound. Chemistry & Biology, 2010, 17(6): 659-664.