Redox Proteomics Service
What is Redox Proteomics?
Redox proteomics is a specialized branch of post-translational modification (PTM) proteomics that systematically investigates redox-dependent changes in protein structure, activity, and interactions. By profiling these changes across the proteome, it helps explain how cells use redox balance to control normal functions and respond to stress.
Because redox modifications are dynamic and often reversible, their analysis relies on high-resolution mass spectrometry, selective chemical labeling, and robust data analysis. These capabilities make redox proteomics a valuable discovery tool in cell biology, pharmaceutical research, environmental studies, and bioengineering.
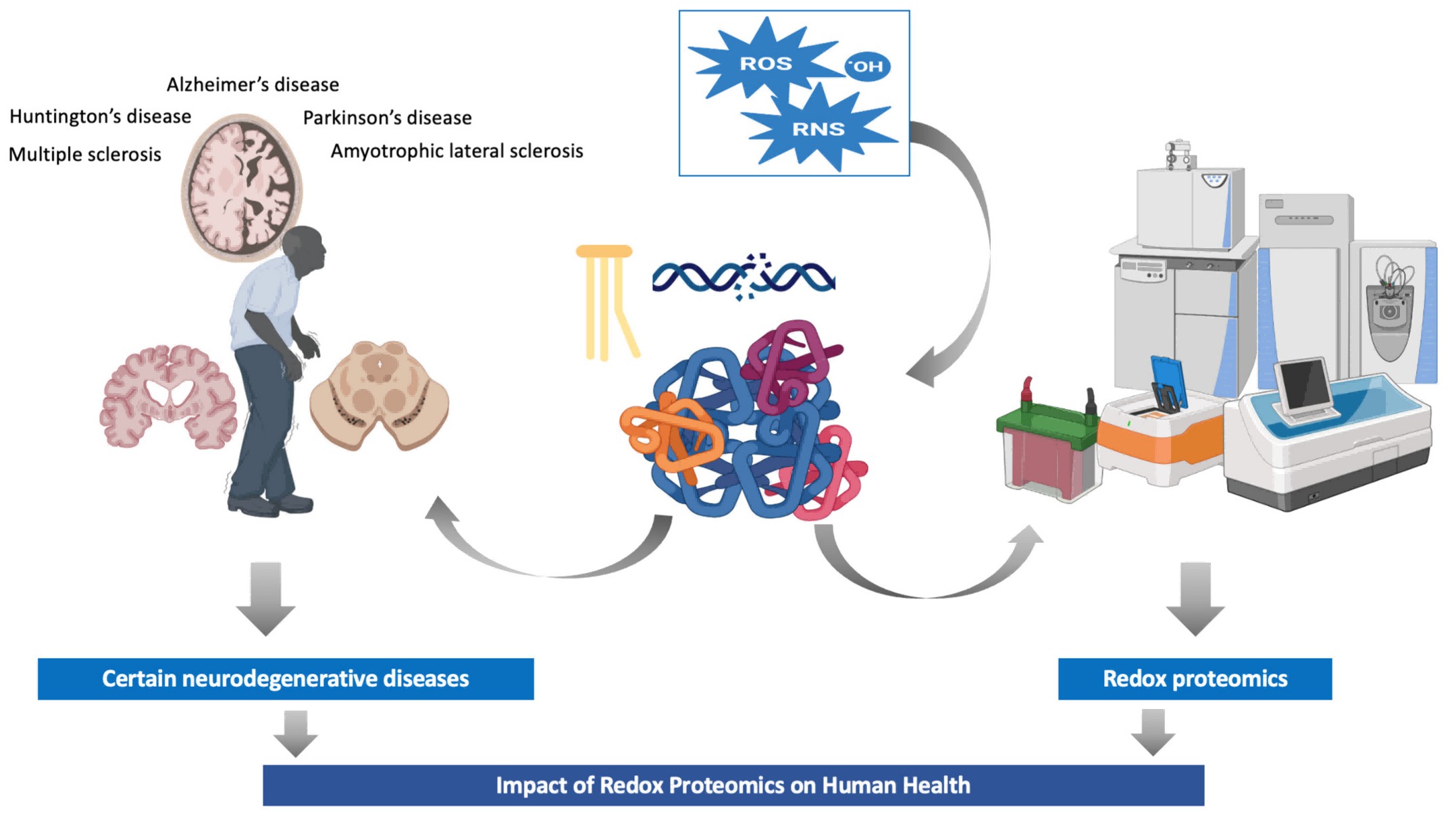
Figure 1. Redox proteomics: impact on human health and new insights (Cadenas-Garrido P, et al., 2024).
Why Redox Proteomics Requires Specialized Analytical Strategies
Conventional proteomics workflows often struggle to capture redox modifications because these modifications are chemically unstable and can be easily altered during sample handling. As a result, crucial oxidation-dependent protein features can be lost or misrepresented.
Preserving Native Redox States: Careful control of oxidation and reduction during sample handling is essential. Antioxidant-controlled preparation minimizes artificial modification while preserving endogenous redox states.
Chemical Labeling and Selective Enrichment: Differential chemical labeling, switch-based assays, and affinity capture strategies stabilize redox modifications and enrich modified peptides. These steps are critical for improving sensitivity and site-localization confidence.
PTMs Analyzed by Redox Proteomics
S-Nitrosylation
S-nitrosylation is a reversible protein modification in which nitric oxide attaches to a cysteine sulfur atom. This small chemical change can switch enzymes on or off, influence how long a protein remains stable, and fine-tune cellular signaling pathways. Using high-resolution redox proteomics, researchers can accurately pinpoint where S-nitrosylation occurs and measure how these modifications change across different biological conditions, providing clear insight into nitric oxide–driven regulation.
S-Glutathionylation
S-glutathionylation is a reversible modification in which glutathione forms a mixed disulfide bond with a protein cysteine. This process acts as a redox buffer, temporarily shielding sensitive cysteine residues from permanent oxidative damage. At the same time, it serves a regulatory role by modulating protein activity as cellular redox conditions change, thereby fine-tuning signaling pathways during oxidative stress and recovery.
Disulfide Bond Formation and Rearrangement
Both intra- and intermolecular disulfide bonds influence protein folding, complex assembly, and structural stability. Redox proteomics captures dynamic disulfide remodeling that cannot be resolved by standard proteomics alone.
Quantitative Redox Proteomics Strategies
Multiplexed Labeling with TMT or iTRAQ
Isobaric labeling techniques, such as TMT or iTRAQ, attach a chemical tag to peptides from different samples. This allows multiple conditions to be analyzed in a single mass spectrometry run.
Label-Free Quantitation for Flexibility
Label-free quantitation relies on measuring peptide signal intensity or counting the number of spectra corresponding to a peptide. While it does not require chemical tagging, it depends on careful experimental design and high-quality sample preparation to maintain consistency.
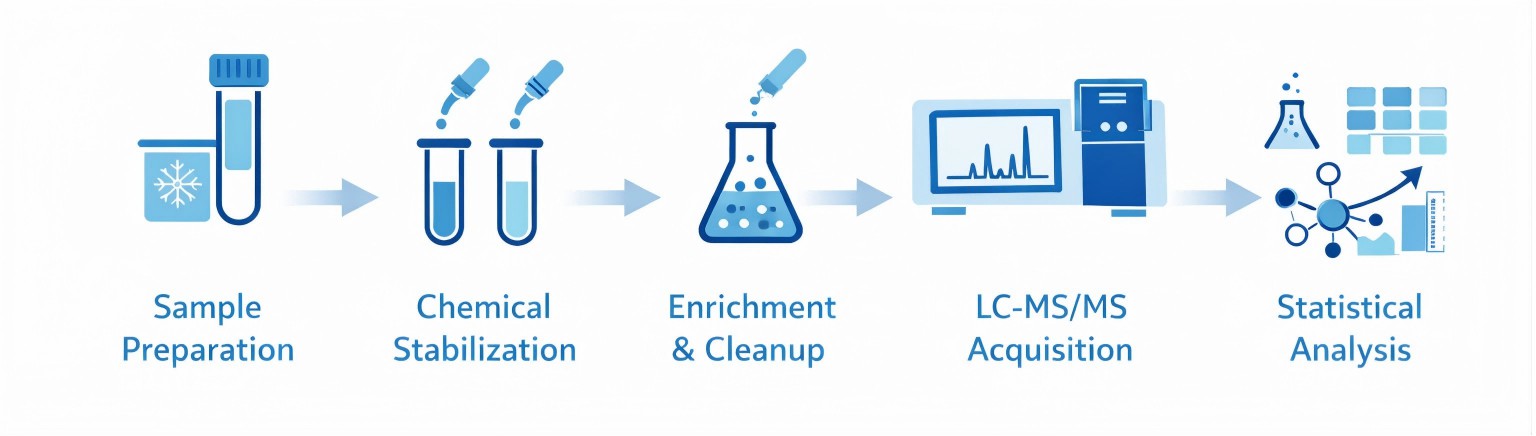
Creative Proteomics' Redox Proteomics Service Workflow
- Sample preparation: Document sample provenance; check integrity; store and process under antioxidant and low-temperature conditions to preserve native redox states.
- Chemical stabilization and differential labeling: Apply switch assays, thiol-blocking, or isotopic labeling to stabilize and mark endogenous redox modifications prior to enrichment.
- Enrichment and cleanup: Use affinity capture, resin-assisted capture, or biotin-switch derivatives to concentrate modified species and reduce background.
- LC-MS/MS acquisition: Optimize acquisition parameters (survey/fragmentation cycles, dynamic exclusion, resolution) to balance depth and quantitation.
- Statistical analysis: Generate volcano plots, heatmaps, motif enrichment analyses, and PPI mapping to contextualize redox changes within pathways.
Deliverables and Reporting Standards
- Comprehensive methods and metadata: Detailed sample handling, reagents, LC and MS settings, and enrichment protocols.
- Peptide- and site-level quantitation tables: CSV/Excel with localization confidence, intensities, ratios, and missing-value indicators.
- Visual reports: Volcano plots, heatmaps, motif enrichments, PPI networks, and pathway summaries.
- Raw data and search files: Vendor raw files, and search engine outputs for independent reanalysis.
- Interpretation summary: Brief expert commentary linking major redox changes to biological processes with suggested follow-up experiments.
Applications of Redox Proteomics in Research and Industry
- Mapping redox signaling in cell biology and stress responses: Identify redox-regulated enzymes and signaling nodes to generate targeted hypotheses.
- Aging and tissue-specific redox remodeling: Profile age-dependent shifts in cysteine oxidation across tissues to study functional decline and resilience.
- Drug mechanism-of-action studies: Identify off-target redox effects or redox-mediated target engagement to inform chemical optimization.
- Environmental toxicology and pollutant impact studies: Use redox signatures to evaluate oxidative stress responses to environmental toxins and pollutants.
- Metabolic engineering and bioprocess optimization: Monitor redox state shifts in engineered strains to improve yield, folding, and stability of bioproducts.
Sample Requirements for Redox Proteomics Analysis
| Sample type | Recommended amount | Preservation & storage | Key notes / constraints |
| Animal tissue (fresh) | 200–500 mg | Snap-freeze; store at −80°C | Minimize dissection time; avoid air exposure |
| Plant tissue (fresh) | 200–500 mg | Snap-freeze; store at −80°C | Protect from light; minimize thaw cycles |
| Suspension cells | >3×10⁷ cells | Pellet quickly, remove media, snap-freeze; −80°C | Quench metabolism; avoid lysis detergents before processing |
| Adherent cells | >3×10⁷ cells | Detach, pellet, snap-freeze; −80°C | Process rapidly to preserve native redox state |
| Microorganisms | ≥50 mg wet or >3×10⁷ cells | Pellet, snap-freeze; −80°C | Remove extracellular oxidants; cold chain for transport |
| Serum / plasma | ≥1 mL | Aliquot; store −80°C (short-term −20°C) | Use anticoagulant-consistent collection; avoid hemolysis |
| Purified protein lysate | Total protein ≥3 mg; ≥1 µg/µL | Aliquot; snap-freeze; −80°C | Add appropriate stabilizers; avoid strong reducing/oxidizing agents |
Why Choose Creative Proteomics for Redox Proteomics Services
- Extensive Expertise: Years of experience in proteomics and redox biology ensures reliable, high-quality analysis.
- Preservation of Native Redox States: Strict sample handling and chemical stabilization prevent artificial oxidation or reduction.
- Targeted Detection: Advanced labeling and enrichment strategies reveal low-abundance oxidative modifications with precision.
- High-Resolution Mass Spectrometry: LC-MS/MS enables confident identification and localization of redox modification sites.
- Robust Data Analysis: Bioinformatics workflows include statistical validation, pathway mapping, and clear visual outputs for easy interpretation.
FAQ
-
Q1: Which amino acid residues are most susceptible to redox modifications?
A1: Cysteine thiols are the primary redox sensors in proteins due to their nucleophilic reactivity, leading to reversible modifications (e.g., disulfide bonds, sulfenic acid) and irreversible oxidation products under stress. Other residues like methionine and tyrosine also undergo oxidation but with different biological implications.
-
Q2: What is the difference between redox proteomics and oxidative stress profiling?
A2: Oxidative stress profiling measures the presence and effects of reactive species, whereas redox proteomics specifically identifies and quantifies oxidative PTMs on proteins to define how redox chemistry alters protein function and regulation.
-
Q3: How are redox proteomics data validated?
A3: Orthogonal methods such as targeted MS (PRM/SRM), mutation analysis, and biochemical assays help confirm redox PTM identification and functional relevance
Demo
Demo: Quantitative analysis of redox proteome reveals oxidation-sensitive protein thiols acting in fundamental processes of developmental hematopoiesis
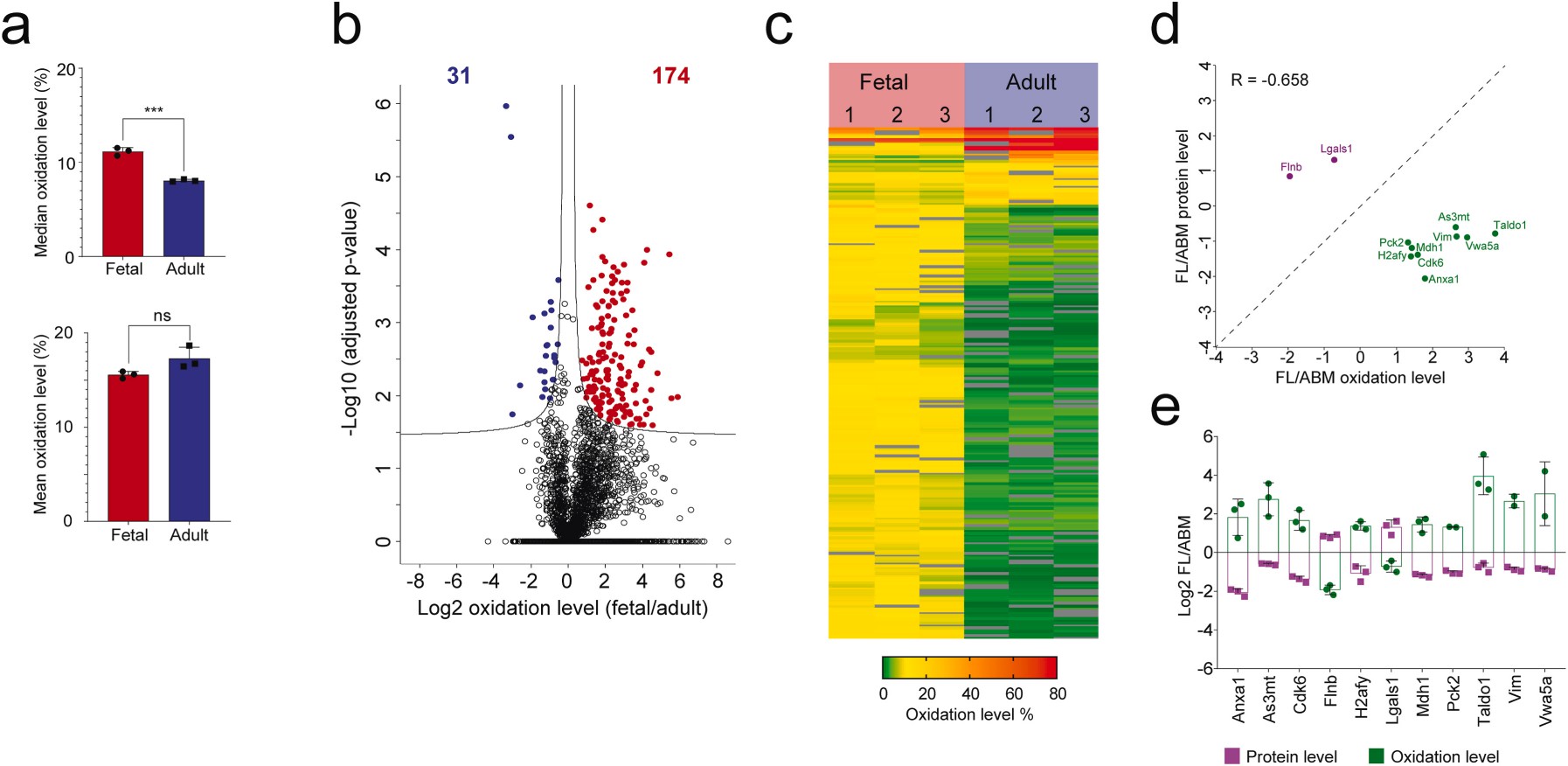
Figure 2. Redox modulation of fetal cysteine proteins in HSPCs (Pimkova K, et al., 2022).
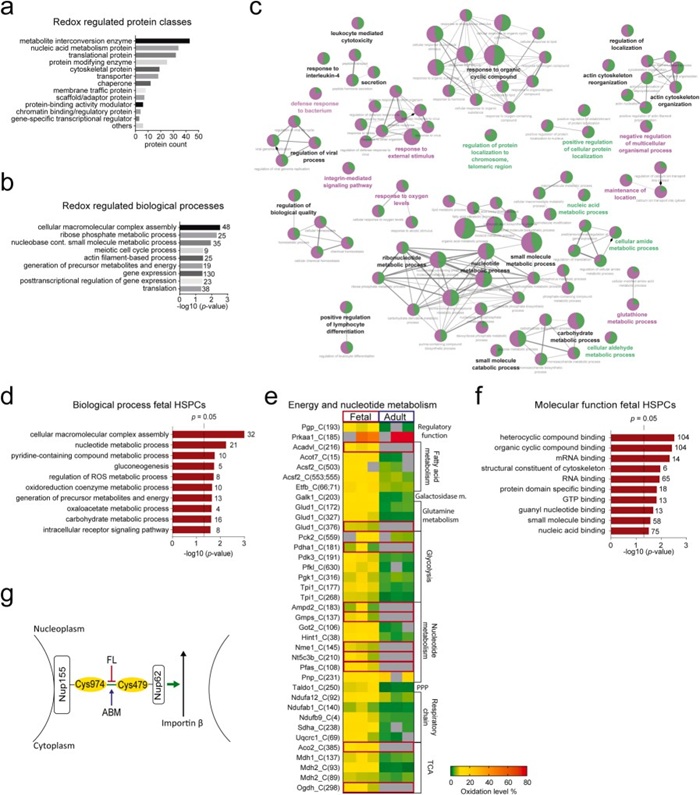
Figure 3. GO enrichment and Functionally grouped annotation network of HSPC proteins (Pimkova K, et al., 2022).
-
Case Study
Case: Global redox proteome and phosphoproteome analysis reveals redox switch in Akt.
Author: Su Z, et al.
Journal: Nature communications
Publication: 2019
DOI: 10.1038/s41467-019-13114-4
Abstract
The study maps proteome-wide reversible cysteine oxidation and the phosphoproteome in adipocytes under chemically induced oxidative stress, revealing extensive crosstalk between cysteine oxidation and phosphorylation signaling. A key finding is that oxidation of two PH-domain cysteines in Akt (C60, C77) forms a redox switch that stabilizes the PIP₃ binding pocket, promotes membrane recruitment and modulates downstream substrate phosphorylation.
Methods
- Redox proteomics: iodoTMT 6-plex labelling of reduced/oxidized cysteines, peptide HILIC fractionation, and LC–MS/MS. Four replicates with forward/reverse labelling were used. Normalization to protein abundance (from SILAC) was applied for 24 h conditions.
- Total proteome: Triple SILAC labelling and LC–MS/MS; protein abundance estimated to control for expression changes.
- Phosphoproteomics: EasyPhos workflow, label-free LC–MS/MS; four biological replicates; >23,000 unique phosphorylation events quantified.
- Targeted IP-MS for Akt: Non-reducing FLAG-Akt2 immunoprecipitation, tryptic digestion, LC–MS/MS, and cross-linked/disulfide searches to map disulfides and glutathionylation.
- Cell and in vivo functional assays: Mutagenesis, overexpression systems including MK2206-resistant Akt2-W80A, western blotting of Akt phosphorylation and substrates, TIRF microscopy for PM translocation, soft-agar growth assays and Drosophila genetic
Results
- Measured ~13,451 reversible cysteine oxidations; 24-h BCNU/AF strongly increased differentially oxidized peptides.
- Identified >23,000 phosphorylation events; oxidative stress altered phospho-signaling primarily by reducing signal fidelity rather than uniformly blocking kinase activation. Example: despite Akt T309 hyper-phosphorylation, phosphorylation of 11/36 Akt substrates was inhibited.
- Akt harbors two reversible modifications: a PH-domain disulfide C60–C77 that enhances PIP₃ binding and PM recruitment, and a kinase-domain disulfide C297–C311 linked to decreased catalytic output.
- The cancer mutation C77F phenocopied the oxidized PH domain. MD and biochemical docking supported that the C60–C77 disulfide stabilizes the PIP₃ pocket. NOX inhibition reduced insulin-stimulated Akt recruitment, consistent with ROS-dependent regulation.
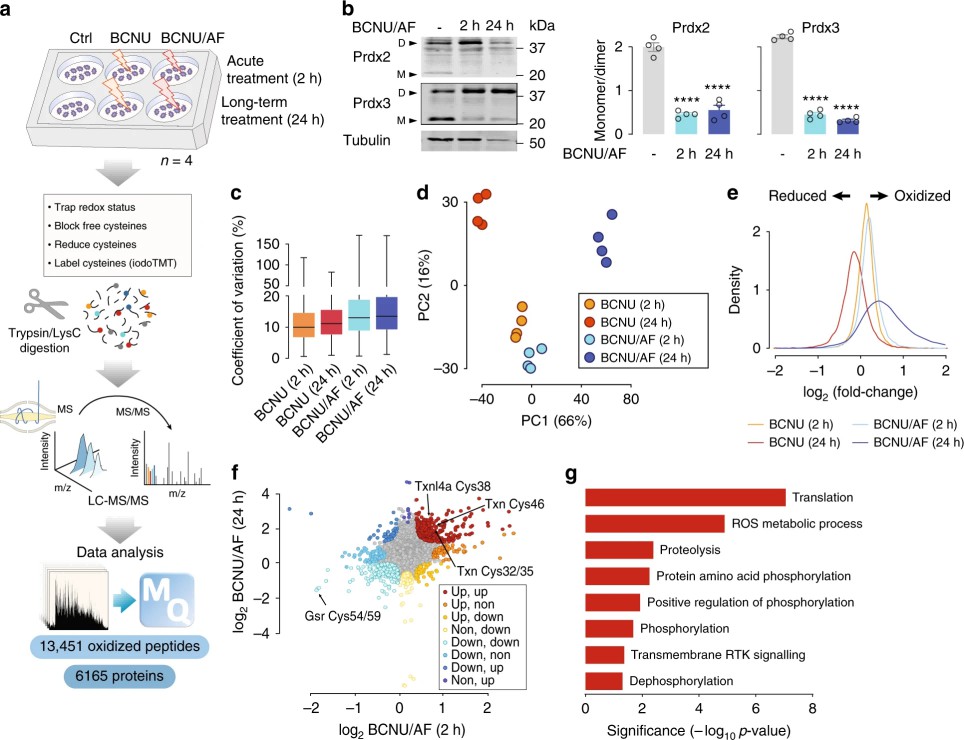
Figure 4. Redox proteomic analysis of adipocytes.
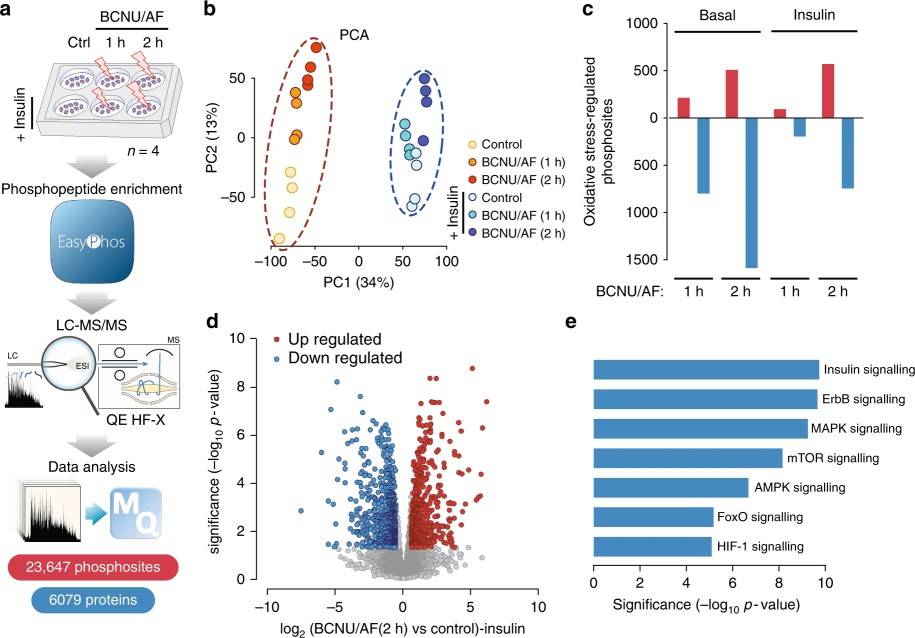
Figure 5. Oxidative stress-regulated phosphoproteome in adipocytes.
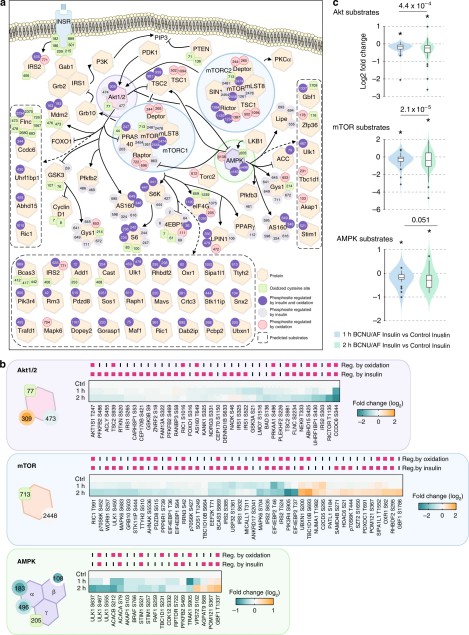
Figure 6. Integrative analysis of oxidative stress-regulated phospho-signalling networks.
Conclusion
This study shows that oxidative stress reshapes cell signaling by changing how selectively kinases act on their targets, rather than by simply increasing or decreasing kinase activity. By combining redox proteomics with phosphorylation analysis, the authors uncover a redox-dependent mechanism that controls Akt movement to the cell membrane through reversible cysteine oxidation. These results emphasize that redox-sensitive protein modifications play a central role in fine-tuning signaling pathways and demonstrate the value of redox proteomics for uncovering signaling interactions at the system level.
Related Services
References
- Cadenas-Garrido P, et al. Using redox proteomics to gain new insights into neurodegenerative disease and protein modification. Antioxidants, 2024, 13(1): 127.
- Su Z, et al. Global redox proteome and phosphoproteome analysis reveals redox switch in Akt. Nature communications, 2019, 10(1): 5486.
- Pimkova K, et al. Quantitative analysis of redox proteome reveals oxidation-sensitive protein thiols acting in fundamental processes of developmental hematopoiesis. Redox Biology, 2022, 53: 102343.