Exposome Microarray Profiling Service
What is Exposome?
The exposome describes the complete environmental exposures a person experiences from conception through life. It includes physical, chemical, biological, and social factors that interact with the body. Unlike the genome, which is largely stable, the exposome is dynamic and changes over time. It covers diet, pollutants, lifestyle habits, and interactions with the microbiome. Studying the exposome provides a broad view of how external influences shape health and disease risk.
Figure 1. The exposome as an analytical framework linking exposures to outcomes (Niedzwiecki M M, et al., 2019).
Why Use Microarray for Exposome Analysis?
The exposome refers to the total environmental exposures and lifestyle factors that shape health over a lifetime. Studying it helps researchers see how these exposures interact with genes and biological regulation to influence disease risk. Exposome microarray profiling is a modern method that uses microarray platforms to measure the biological impact of environmental factors. In a single test, it can detect proteins, metabolites, and gene activity changes linked to chemical or physical stress.
Unlike traditional methods that measure one target at a time, microarray profiling uses thousands of probes on a small chip to capture many signals simultaneously. This generates large, detailed datasets that connect exposure patterns to disease processes and potential therapeutic targets.
Differences Between Exposome Microarray Profiling and Standard Microarray Assays
| Feature | Exposome Microarray Profiling | Standard Microarray Assays |
| Primary Objective | Assess environmental exposures' influence on molecular pathways and disease risk. | Measure expression levels of predefined genes or proteins. |
| Scope of Biomarkers | Broad coverage, including proteins, metabolites, epigenetic marks, and environmental signatures. | Limited to specific genes, transcripts, or proteins. |
| Study Design Focus | Integrates environmental, genetic, and phenotypic data for holistic exposome studies. | Focuses on analyzing a specific biological pathway or cellular process. |
| Applications | Environmental health, biomarker discovery, population-level exposure analysis, and personalized medicine. | Transcriptomics, targeted proteomics, and pathway-specific studies. |
| Data Interpretation | Requires complex integration of exposure-related data with multi-omics datasets. | Primarily relies on differential expression or signal intensity comparisons. |
| Throughput and Scale | Optimized for high-throughput exposome-wide biomarker profiling across large populations. | Moderate throughput, optimized for smaller-scale experimental studies. |
Exposome Microarray vs. Other Profiling Techniques
Several technologies are available for studying environmental exposures, but each has strengths and limitations.
| Feature | Exposome Microarray | RNA-Seq | LC-MS Proteomics | ELISA |
| Throughput | Very high | High | Moderate | Low |
| Biomarker Coverage | Thousands simultaneously | Genome-wide | Hundreds per run | Single target |
| Cost per Sample | Low to moderate | High | High | Low |
| Data Complexity | Moderate | High | High | Low |
| Best Use Case | Broad exposome profiling | Transcriptome mapping | Protein-specific studies | Target validation |
Advantages of Our Exposome Microarray Profiling Service
Creative Proteomics offers a platform optimized for sensitivity, scalability, and reproducibility. The key benefits include:
- High-Throughput Analysis: Detects thousands of biomarkers in a single run.
- Comprehensive Coverage: Simultaneously profiles proteins, metabolites, and gene expression.
- Cost-Effectiveness: Reduces per-sample analysis costs compared to sequencing-based methods.
- Scalability: Suitable for individual studies and large population-based projects.
- Standardized Quality Control: Ensures data consistency and comparability across experiments.
Workflow for Exposome Microarray Profiling Service
Creative Proteomics applies a standardized yet highly adaptable workflow to ensure reproducible and accurate profiling results. The process involves several critical steps:
- Sample Preparation: High-quality biological samples, such as serum, plasma, urine, or tissue lysates, are collected and processed. Rigorous purification methods ensure minimal background interference.
- Microarray Hybridization: Purified samples are labeled with fluorescent or chemiluminescent tags and hybridized onto a microarray chip containing thousands of probes. Each probe corresponds to a biomarker associated with environmental exposure or physiological response.
- Signal Detection and Quantification: Advanced scanners capture high-sensitivity hybridization signals. Signal intensity correlates with biomarker abundance, providing quantitative insights into molecular alterations.
- Bioinformatics Analysis: Comprehensive statistical and pathway analyses identify differentially expressed biomarkers and associate them with specific exposure patterns, disease states, or drug responses.
Deliverables and Reporting Standards
Creative Proteomics integrates advanced bioinformatics workflows to deliver reliable and publication-ready datasets. Key components include:
- Normalization: Reducing technical variability across microarray chips.
- Statistical Modeling: Identifying significant biomarkers linked to exposures or disease.
- Pathway Mapping: Connecting altered biomarkers to biological pathways.
- Data Visualization: Delivering interpretable results using heatmaps, volcano plots, and network diagrams.
Applications of Exposome Microarray Profiling
- Biomarker Discovery: Identifying signatures associated with environmental exposures, disease onset, and therapeutic responses.
- Toxicology Studies: Exposome microarrays provide insights into toxicity pathways by measuring how cells or tissues respond to environmental chemicals. This is particularly useful for evaluating industrial pollutants, food additives, or drug by-products.
- Personalized Medicine: Every individual has a unique combination of genetic traits and environmental exposures. Exposome profiling helps capture this complexity by showing how external factors influence biological responses. This information supports the development of tailored treatment strategies, ensuring that therapies are both safer and more effective.
- Pharmaceutical Development: Drug candidates can behave differently depending on environmental context. Microarray profiling helps drug developers understand how exposures such as diet, smoking, or stress influence drug metabolism and toxicity.
- Epidemiological Research: On a population level, exposome profiling can reveal patterns of exposure linked to disease prevalence. For instance, comparing biomarker profiles across communities with different lifestyles or pollution levels can clarify how environmental factors drive public health outcomes.
Simple Requirements
| Sample Types | Serum, plasma, urine, tissue lysates, or cell extracts |
| Minimum Volume | 200–500 µL for fluids; 20–50 mg for tissues |
| Storage Conditions | Store at –80 °C; avoid repeated freeze–thaw cycles |
| Shipping Requirements | Ship on dry ice in leak-proof containers; include detailed sample metadata |
| Quality Criteria | Samples should be free of hemolysis, contamination, and degradation |
Why Choose Creative Proteomics for Exposome Microarray Profiling
Creative Proteomics has over two decades of experience in delivering high-precision omics-based solutions for the biomedical community. Our exposome microarray profiling platform is supported by:
- State-of-the-art microarray technologies
- Skilled bioinformatics experts
- Comprehensive multi-omics integration capabilities
- Customizable experimental workflows
- Proven success in supporting academic, clinical, and industrial research
FAQ
-
Q1: Can exposome microarray profiling detect unknown environmental exposures?
A1: While microarray platforms primarily focus on known biomarkers, Creative Proteomics combines custom array design with integrated LC-MS proteomics to enable expanded biomarker discovery. By incorporating multi-omics data, we can identify novel exposure signatures that may not yet be included in commercial panels.
-
Q2: What controls and replicates are necessary?
A2: The study design should include negative controls, positive controls, and technical replicates. The laboratory provides spike-in controls when appropriate.
-
Q3: What validation strategies are recommended for discovery findings?
A3: Orthogonal validation using LC-MS, qPCR, or ELISA is recommended. The laboratory can perform targeted validation upon request.
-
Q4: Can the microarray be customized for novel biomarkers?
A4: Yes. Custom probe sets and bespoke panels are available. The laboratory supports target selection and assay validation.
-
Q5: How does exposome microarray profiling differ from standard microarray assays?
A5: Exposome microarray profiling leverages microarray platforms to measure biomarkers specifically related to environmental exposures. Unlike traditional gene-expression microarrays, it may include probes for exposure markers, proteins, metabolites, or adducts, enabling simultaneous, multiplexed detection of exposure-related signatures.
Demo
Demo: MicroRNA profile for health risk assessment: miRNA changes in response to persistent organic pollutants (POPs).
The study used high-throughput microarray profiling of microRNAs to identify miRNA signatures associated with POP exposure in a population cohort. The authors associated altered miRNA expression with exposure metrics and suggested biomarker potential.
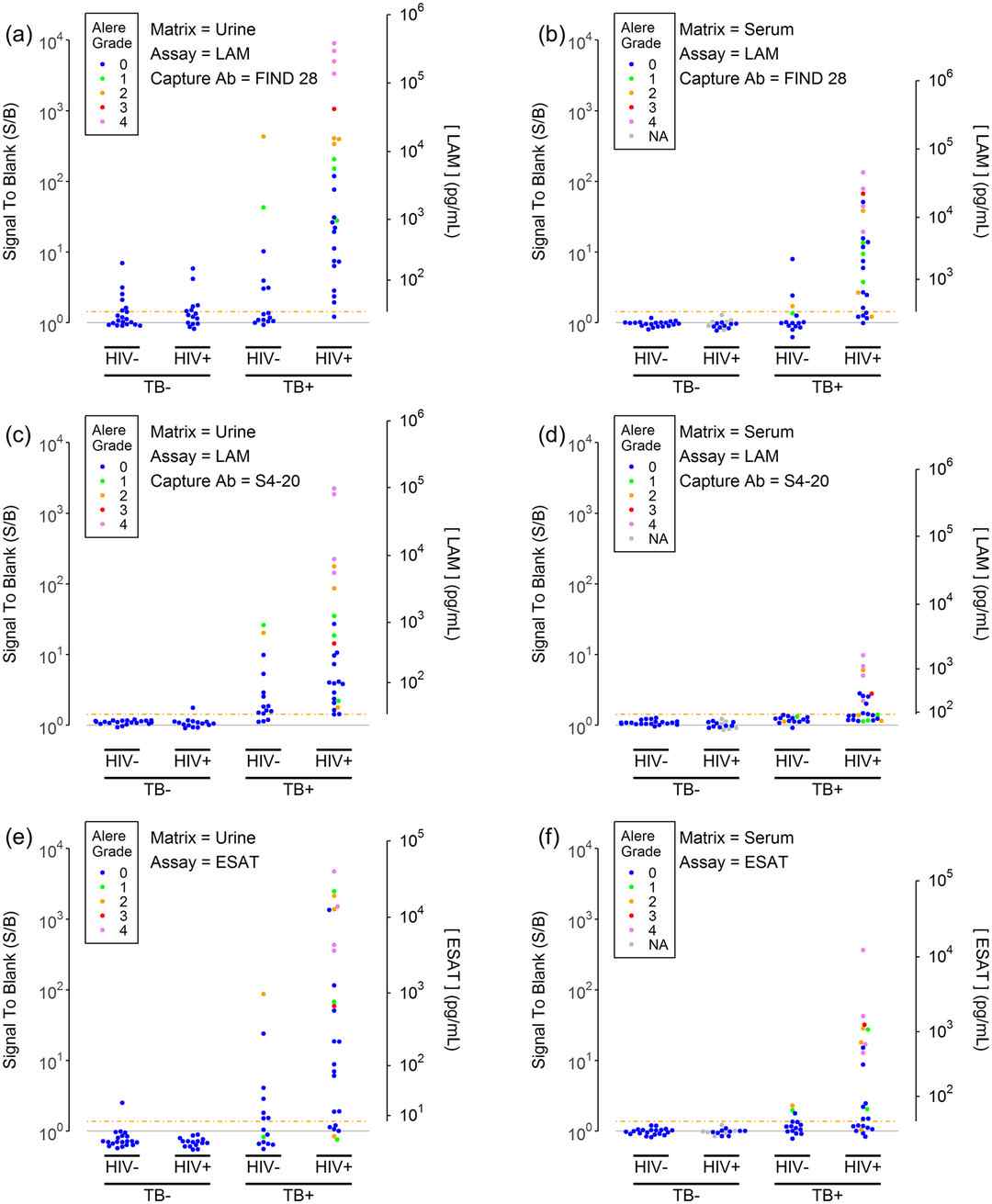
Figure 2. Expression levels of the top 12 associated miRNAs in the defined exposure groups. (Krauskopf J, et al., 2017).
-
Case Study
Case: Blood and urine multi-omics analysis of the impact of e-vaping, smoking, and cessation: from exposome to molecular responses
Abstract:
Tobacco harm reduction (THR) aims to reduce health risks by encouraging smokers to switch from combustible cigarettes to less harmful nicotine delivery products. Assessing THR effects is challenging because smokers may not fully switch, resulting in heterogeneous exposure patterns. Understanding these patterns requires linking chemical exposures to biological effects. This study aimed to (i) characterize individual exposomes, including biomarkers of exposure (BoE) and broader molecular profiles, and (ii) assess biological responses across systemic compartments in cigarette smokers (CS), e-vapor users (EV), former smokers (FS), and never smokers (NS).
Methods
- Exposome assessment: Targeted BoE analysis in urine (e.g., cotinine, CO, CEMA, tNNAL), and untargeted metabolomics for plasma and urine to capture additional exposures (e.g., THC, drugs).
- Microarray/transcriptomics: RNA from isolated blood cell types (T4, T8, B cells, monocytes, neutrophils, platelets, RBCs) and whole blood analyzed via microarrays to identify gene expression changes linked to exposures.
- Other omics: Proteomics, lipidomics, and DNA methylation profiling.
- Data analysis: Multivariate analyses, machine learning models to predict smoking status and link exposures to biological effects.
Results
- Exposome characterization: PCA-based combustion and nicotine scores distinguished CS, EV, FS, and NS, revealing heterogeneity in EV and FS groups, often due to dual use or incomplete cessation. Untargeted metabolomics detected additional exposures, such as THC and pharmaceuticals.
- Microarray/transcriptomics findings: Differential gene expression reflected smoking-related biological effects, particularly in T4, T8, B cells, and monocytes. Key genes (e.g., GPR15, AHRR, CLDND1) were regulated by xenobiotic sensors like AHR. Transcriptomics provided high predictive performance (MCC 0.6–0.9) for smoking status.
- Other omics: Proteomics and lipidomics captured downstream biological responses (e.g., inflammation), with plasma proteomics showing intermediate predictive performance. DNA methylation changes were prominent in CS and largely reversible in FS and EV.
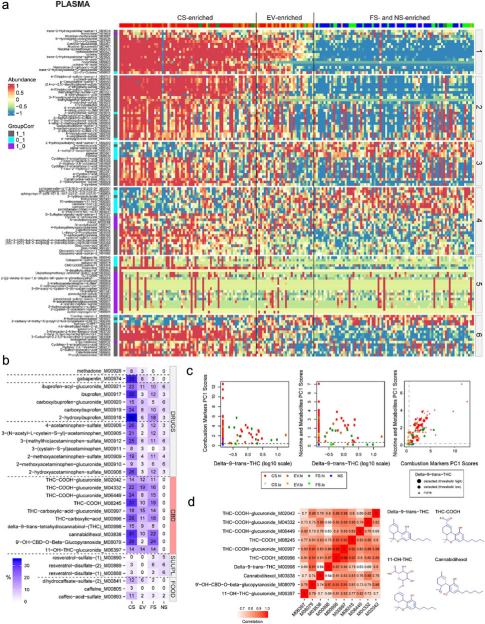
Figure 3. Identification of potential novel BoE and deeper subject exposome investigations in plasma and urine using untargeted metabolomics.
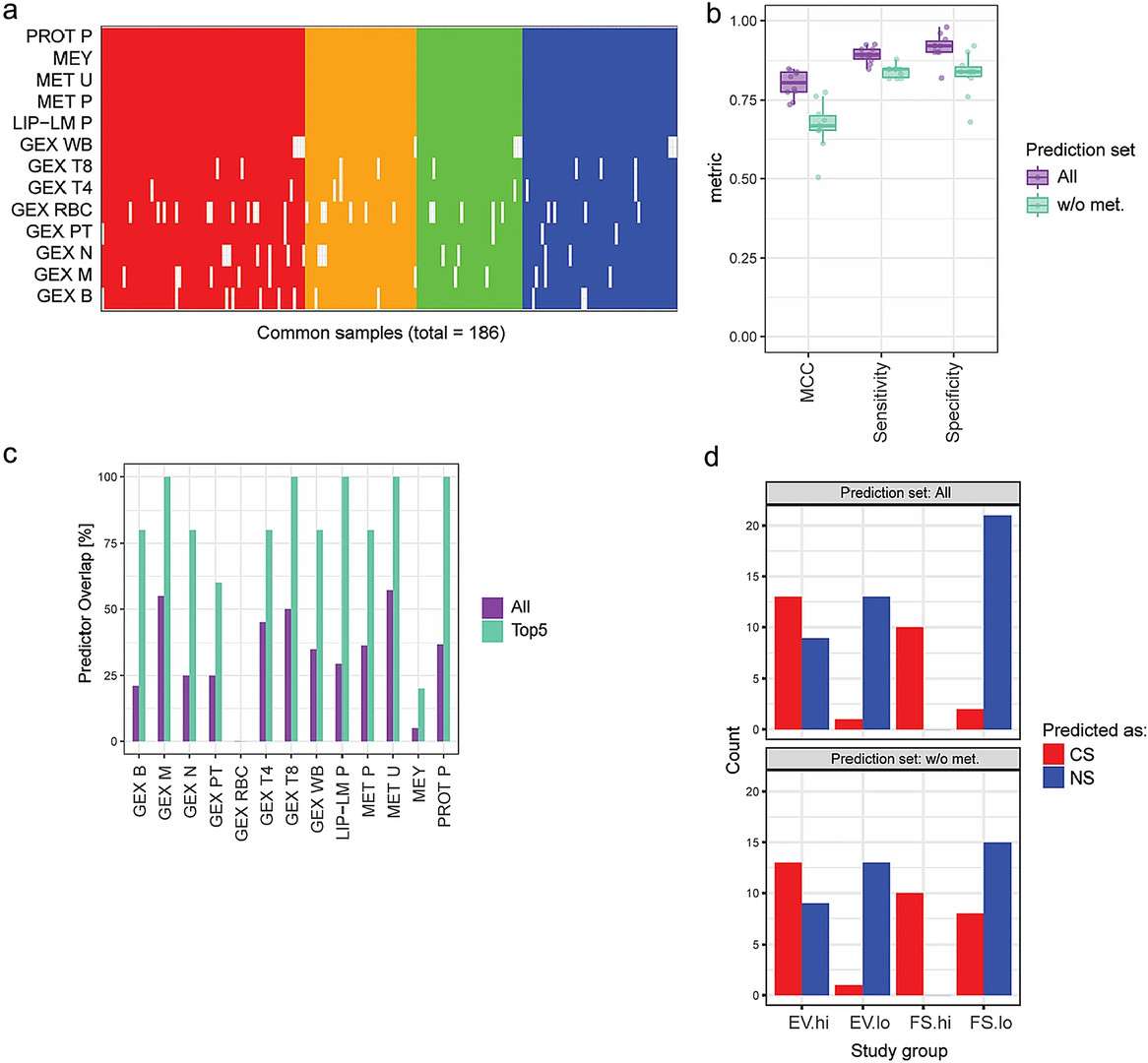
Figure 4. Integrated across-omics model and associated molecular signatures predictive of smoking status.
Conclusion
This comprehensive exposome-omics analysis demonstrates that early-life exposures leave discernible molecular imprints in childhood across multiple biological layers. The study furnishes a public catalogue of exposure-omics associations to catalyse further mechanistic and translational research. The results underscore the value of integrating microarray-based and targeted omics platforms in exposome research.
Related Services
References
- Niedzwiecki M M, et al. The exposome: molecules to populations. Annual review of pharmacology and toxicology, 2019, 59(2019): 107-127.
- Krauskopf J, et al. MicroRNA profile for health risk assessment: Environmental exposure to persistent organic pollutants strongly affects the human blood microRNA machinery. Scientific reports, 2017, 7(1): 9262.
- Cui Y, et al. The exposome: embracing the complexity for discovery in environmental health. Environmental health perspectives, 2016, 124(8): A137-A140.
- Poussin C, et al. Blood and urine multi-omics analysis of the impact of e-vaping, smoking, and cessation: from exposome to molecular responses. Scientific Reports, 2024, 14(1): 4286.

