Exosome Proteomics
What Is Exosome Proteomics?
Exosome proteomics is the systematic analysis of proteins carried by exosomes, small extracellular vesicles secreted by cells into biological fluids. Exosomes play a crucial role in intercellular communication, carrying a rich molecular cargo that includes proteins, nucleic acids, lipids, and metabolites. By profiling exosomal proteins, researchers can gain insights into cellular physiology, pathological states, and disease progression.
Exosome proteomics utilises advanced mass spectrometry–based detection techniques, enabling both qualitative and quantitative profiling of proteins. With rich experience, Creative Proteomics offers high-quality exosome proteomics services, providing valuable insights into your protein research.
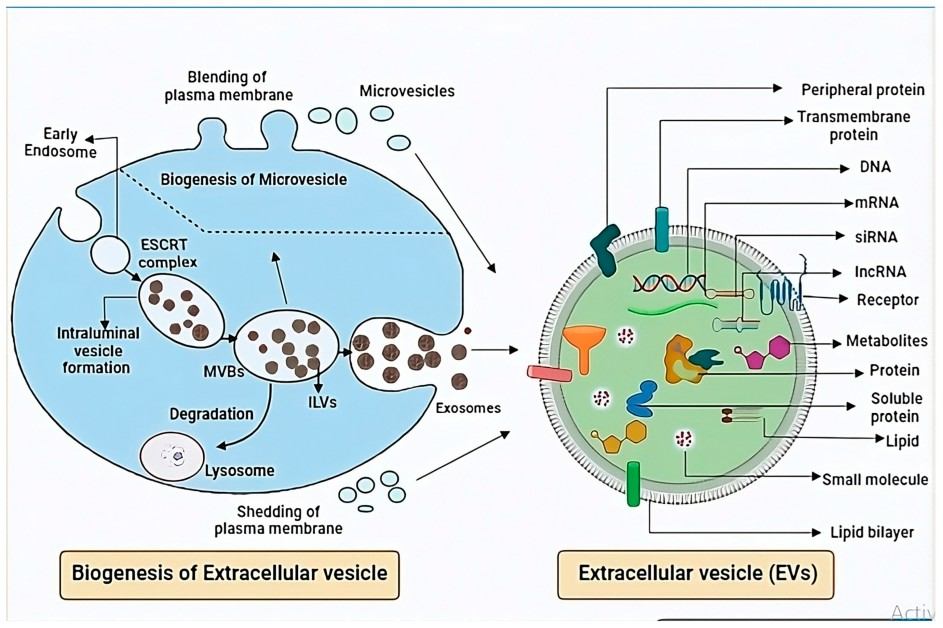
Figure 1. Exosome biogenesis involves multiple stages (Singh M, et al. 2025).
Why Exosomes Are Targets for Proteomic Detection
Exosomes are found in nearly all biological fluids, including blood, urine, saliva, and cerebrospinal fluid. Their small size (30–150 nm) and lipid bilayer structure allow them to transport proteins, nucleic acids, and metabolites stably between cells. This makes them excellent candidates for the discovery of non-invasive biomarkers.
Because exosomes reflect the physiological and pathological state of their parent cells, analyzing their proteomes can reveal disease-specific signatures, including cancer markers, neurodegenerative disease indicators, or immune-modulating proteins. In addition, exosomes provide access to molecular information that is difficult to obtain from tissue biopsies or other cell-derived samples, enabling real-time, minimally invasive disease monitoring.
Proteomic Detection Techniques for Exosomes
LC–MS/MS: Peptides (protein fragments) are separated by tiny-scale chromatography, then measured by a mass spectrometer. Good chromatography increases the number of proteins that can be detected and reduces interference from contaminating proteins.
DIA/4D-DIA: DIA systematically fragments all peptides across defined mass ranges, so it captures low-abundance peptides more reliably across many samples. DIA gives more reproducible quantification for cohort studies and is often our default for biomarker discovery. Adding ion-mobility is like providing the mass spectrometer with another way to separate ions before measurement. This reduces overlap between peptides, increases depth, and speeds up throughput — a valuable feature when sample complexity is high or when both depth and speed are required.
Targeted proteomics (MRM/PRM): For hypothesis-driven studies or biomarker verification, we offer targeted proteomics using PRM or MRM methods. These techniques focus on predefined proteins and measure them with high sensitivity and precision. They are ideal for confirming candidate exosomal markers identified in discovery studies and for supporting downstream functional research.
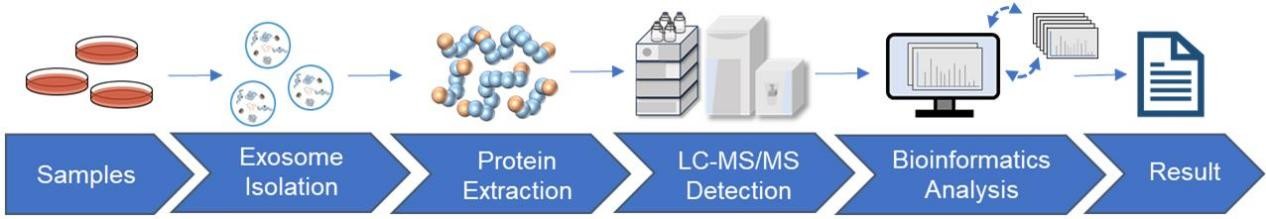
Standardized Workflow for Exosome Proteomics
- Exosome isolation and characterization: Using ultracentrifugation, size-exclusion chromatography, or commercial kits, we isolate high-purity exosomes. Characterization is performed via TEM, NTA, and immunoblotting.
- Protein extraction and enzymatic digestion: Proteins are extracted and digested into peptides, with desalting and concentration to optimize MS detection.
- Mass spectrometry analysis: Samples are analyzed using LC-MS/MS, DIA, or 4D-DIA platforms to generate high-resolution proteome profiles.
- Bioinformatics analysis: Data are processed to identify proteins, quantify relative abundance, annotate functions, and perform pathway and network analyses.
- Results interpretation: Comprehensive reports provide raw data, statistical analyses, and biologically meaningful insights tailored to project goals.
Deliverables and Reporting Standards
- Raw mass spectrometry files
- Processed protein identification and quantification tables
- Statistical and functional analysis results
- Visualizations such as heatmaps, volcano plots, and interaction networks
- Interpretative reports summarizing key findings in a structured format
- PTM-specific datasets (if applicable)
Exosome Proteomics in Biomedical Research
- Cancer Research: Exosomal proteins can serve as early tumor markers, offering minimally invasive diagnostics and enabling real-time monitoring of disease progression. Exosomal proteomic profiles may also reveal mechanisms of therapeutic resistance.
- Neurodegenerative Disease Studies: Exosomes in cerebrospinal fluid, plasma reflect, or urine pathological changes in Alzheimer's, Parkinson's, and other neurodegenerative conditions, providing potential biomarkers for early diagnosis and progression tracking.
- Immunology and Inflammation: Exosomal proteins mediate immune signaling, modulate inflammatory responses, and influence autoimmune processes. Proteomic profiling can elucidate mechanisms of immune regulation and help identify therapeutic targets.
- Drug Development: Exosomes can act as carriers of therapeutic molecules or as targets for drug delivery strategies. Proteomic analysis facilitates assessment of drug efficacy, off-target effects, and molecular interactions in preclinical and clinical studies.
Sample Requirements for Exosome Proteomics
| Plasma / Serum | ≥ 200 μL | ≥ 1×10¹⁰ particles/mL (NTA-based) |
| Urine | ≥ 5–10 mL | Concentration dependent on donor variability |
| Cerebrospinal Fluid | ≥ 200 μL | As available; low-input workflows applicable |
| Cell Culture Supernatant | ≥ 10–50 mL | Dependent on cell density and culture conditions |
| Isolated Exosomes | ≥ 50–100 μg total protein or ≥ 0.25 μg/μL | BCA-quantified protein input |
Why Choose Creative Proteomics for Exosome Proteomics Services
- Expertise in low-input exosome proteomics workflows
- Advanced instrumentation and 4D mass spectrometry capabilities
- DIA and targeted proteomics experience for reproducible quantification
- PTM analysis to expand biological insights
- End-to-end service from isolation to data interpretation
FAQ
-
Q1: How do different exosome isolation methods impact proteomic results?
A1: Different isolation methods (e.g., ultracentrifugation, size‑exclusion chromatography, immunoaffinity capture) influence the purity, yield, and proteome composition of exosome samples. High-purity methods, such as size-exclusion chromatography, reduce the co-isolation of proteins and lipoproteins that can confound downstream mass spectrometry. In contrast, immunoaffinity can selectively enrich exosome subpopulations based on surface markers. Balancing purity and yield is crucial because differences in isolation can result in distinct proteome profiles, which in turn affect biomarker discovery and reproducibility.
-
Q2: What controls should be included in exosome proteomics experiments?
A2: Robust controls include non‑exosomal protein controls, spike‑in standards for quantification, and technical replicates. Additionally, particle characterization (e.g., nanoparticle tracking analysis, electron microscopy) and marker verification (e.g., CD9, CD63) help confirm exosome identity and quality before proteomic analysis begins.
-
Q3: Can exosome proteomic profiles reflect the tissue of origin?
A3: Yes. Because exosomes are secreted by cells and carry cell-specific proteins, their proteomic profiles can reflect the origin of the tissue or disease from which they are derived. For example, serum-derived exosomes have been shown to contain placenta-associated proteins in pre-eclampsia, supporting the utility of exosome proteomics for identifying tissue-specific biomarkers.
-
Q4: Why is standardisation important in exosome proteomics?
A4: Standardisation ensures reproducibility and comparability across studies. Without standardised exosome isolates, data interpretation becomes less reliable, and results may not be generalisable. Standard reference materials and consensus protocols help validate the efficiency of exosome isolation, sample purity, and proteomic detection sensitivity, thereby improving both scientific and clinical utility.
-
Q5: How does protein abundance affect exosome proteomic sensitivity?
A5: Proteomic detection sensitivity can be limited by low protein concentrations in exosome samples. Advanced mass spectrometry techniques and optimised sample preparation can enhance the detection of low-abundance proteins; however, limited input material and heterogeneity remain significant analytical challenges.
Demo
Demo: Comprehensive Proteomic Profiling of Urinary Exosomes and Identification of Potential Non-invasive Early Biomarkers of Alzheimer's Disease in 5XFAD Mouse Model
The authors isolated EVs from human Alzheimer's disease (AD) brain tissue and applied quantitative proteomics to compare AD vs control EV protein cargo. Their analysis identified disease-associated proteomic alterations in brain-derived EVs and used machine-learning approaches to prioritize candidate markers, demonstrating the translational value of tissue-derived EV proteomics.
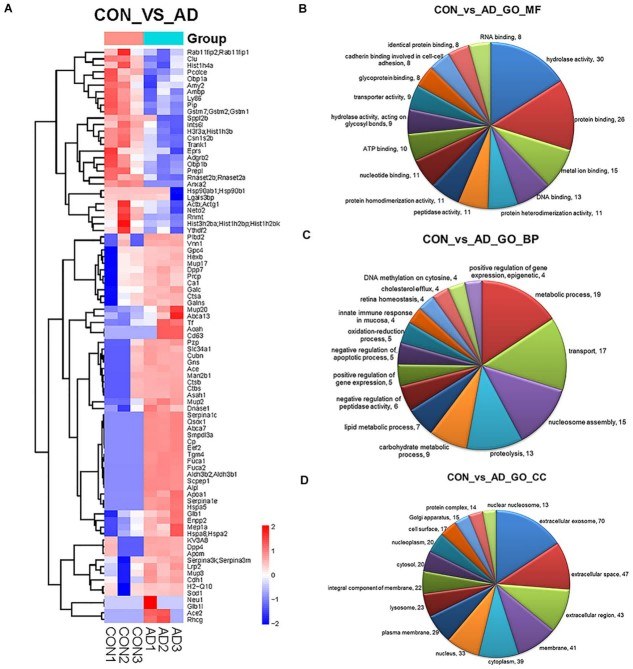
Figure 2. Comparison of the proteomics analysis by hierarchical cluster and gene ontology (Song Z, et al., 2020).
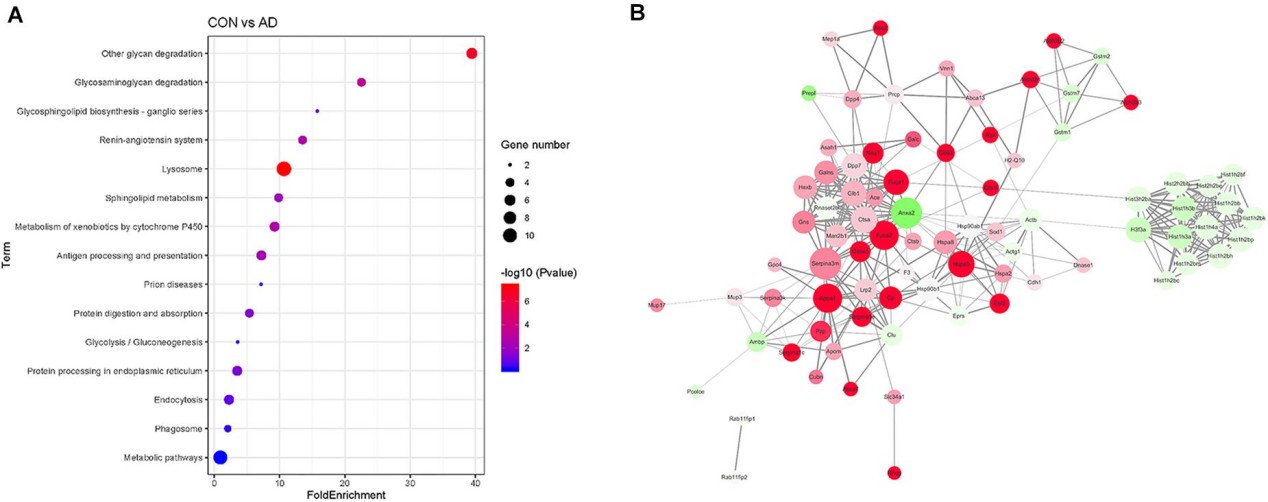
Figure 3. Comparison of the proteomics analysis by KEGG pathway and PPI network analysis
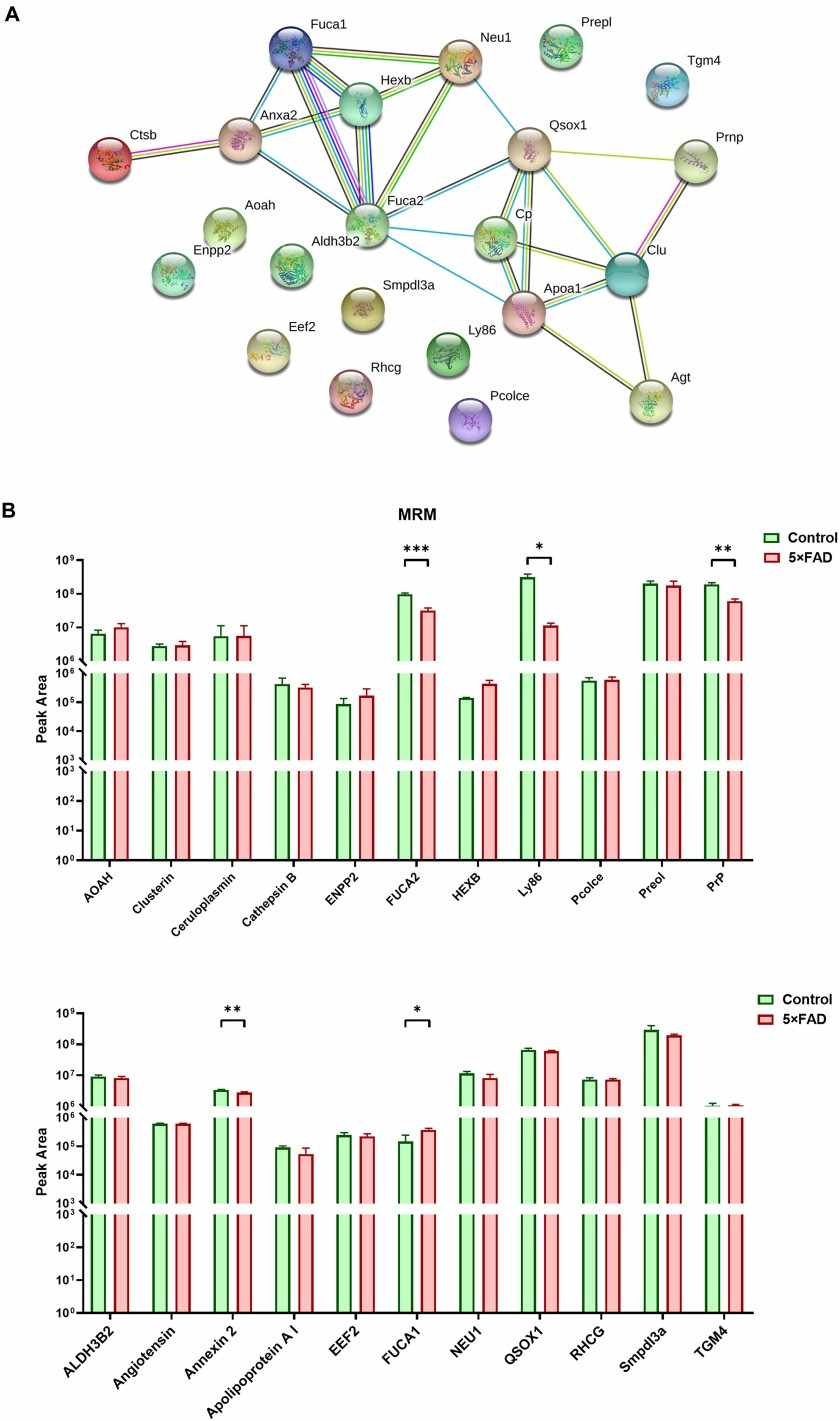
Figure 4. PPI analysis the target proteins from control and 5XFAD mice (Song Z, et al., 2020).
-
Case Study
Case: Isolation and proteomic profiling of urinary exosomes from patients with colorectal cancer
Background
Exosomes are extracellular vesicles that reflect the physiological state of their cells of origin and carry protein cargo with diagnostic potential. Urinary exosomes are especially attractive for noninvasive cancer biomarker discovery because urine can be collected easily and repeatedly, and carries exosomes shed by tumor tissues. However, the proteomic profiles of urinary exosomes in patients with colorectal cancer (CRC) remain underexplored.
Purpose
This study aimed to isolate urinary exosomes from CRC patients and healthy controls and to characterize their protein contents using label‑free liquid chromatography–tandem mass spectrometry (LC–MS/MS). The goal was to reveal differential exosomal protein expression patterns associated with CRC and identify potential protein biomarkers for disease detection and staging.
Methods
- Sample Collection: Urine samples were obtained from CRC patients at different clinical stages and healthy controls.
- Exosome Isolation: Density gradient ultracentrifugation was used to isolate high‑purity urinary exosomes, whose morphology and size (~30–100 nm) were confirmed by electron microscopy and exosomal markers (e.g., CD9, CD63, CD81).
- Proteomic Analysis: Exosomal proteins were extracted and analyzed using label‑free LC–MS/MS for high‑sensitivity protein identification and quantification.
- Statistical and Bioinformatic Processing: Differential protein expression was assessed across samples and correlated with clinical staging.
Results
- The study successfully isolated urinary exosomes from both CRC patients and healthy controls and generated high‑quality proteomic profiles using LC–MS/MS.
- Several exosomal proteins showed statistically significant differential abundance between CRC and control groups, and some varied across clinical disease stages.
- Differentially expressed proteins were associated with pathways relevant to tumorigenesis and CRC progression, suggesting disease‑specific biological signatures in urinary exosomes.
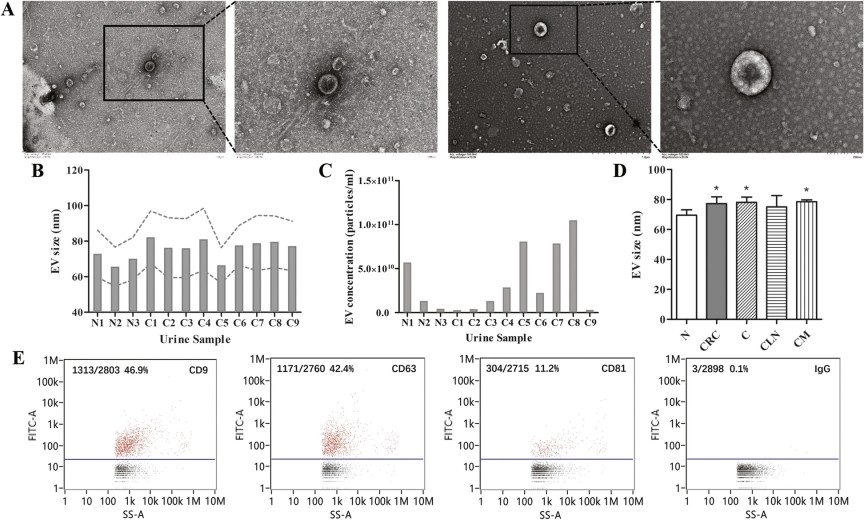
Figure 5. The characterization of EVs isolated from urine.
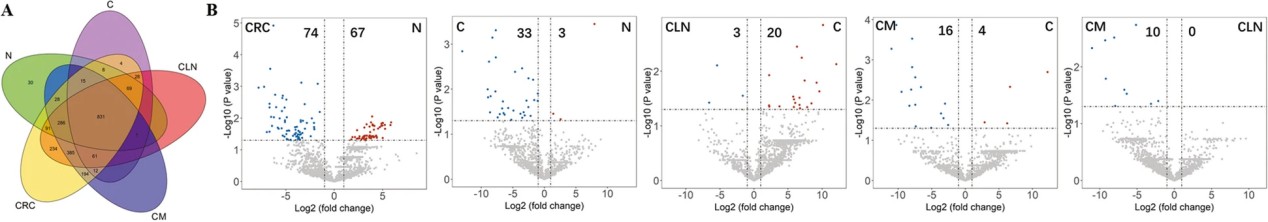
Figure 6. Proteomic profiling and comparison of urinary exosomes from different CRC groups determined by LC-MS/MS.
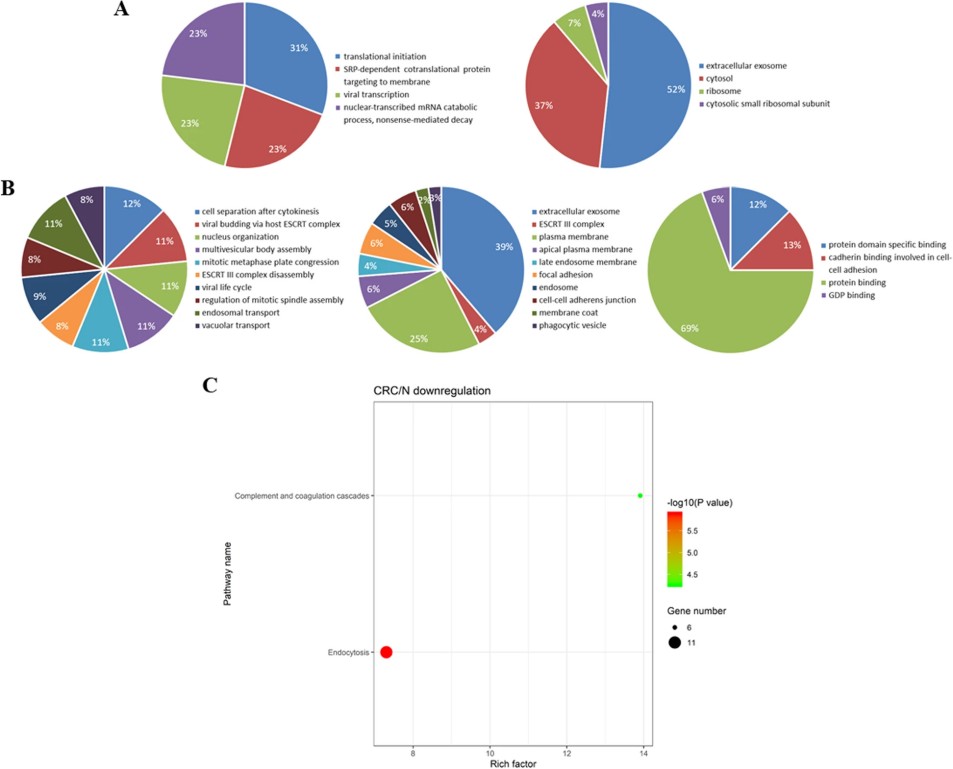
Figure 7. Bioinformatics analysis of differentially expressed proteins identified from the urinary exosomes between the CRC and NC groups.
Conclusion
The proteomic profiling of urinary exosomes revealed distinct protein expression patterns in CRC patients compared to healthy individuals, with stage‑associated changes. These findings demonstrate that urinary exosome proteomics can identify candidate biomarkers for colorectal cancer detection and staging, supporting their potential use in non-invasive diagnostics and personalized monitoring strategies.
Related Services
References
- Singh M, et al. Proteomics of Extracellular Vesicles: Recent Updates, Challenges and Limitations. Proteomes, 2025, 13(1): 12.
- Song Z, et al. Comprehensive proteomic profiling of urinary exosomes and identification of potential non-invasive early biomarkers of Alzheimer's disease in 5XFAD mouse model. Frontiers in genetics, 2020, 11: 565479.
- Ma L, et al. Isolation and proteomic profiling of urinary exosomes from patients with colorectal cancer. Proteome Science, 2023, 21(1): 3.


