Integrative Analysis Services of Proteomics-Acetyl
What is Proteomics-Acetyl Integrative Analysis?
Acetylation involves the covalent addition of an acetyl group (-COCH3) to lysine residues of target proteins, primarily mediated by lysine acetyltransferases (KATs) and reversed by lysine deacetylases (KDACs). This PTM is key in epigenetic regulation, enzyme activity modulation, signal transduction, and protein-protein interaction networks.
Integrative acetyl-proteomics analysis combines quantitative acetylation profiling with other omics datasets to gain deeper biological insights. By linking acetylation changes to transcript levels, metabolic fluxes, or phosphorylation dynamics, researchers can uncover regulatory mechanisms and functional pathways that are otherwise difficult to detect using single-layer analysis.
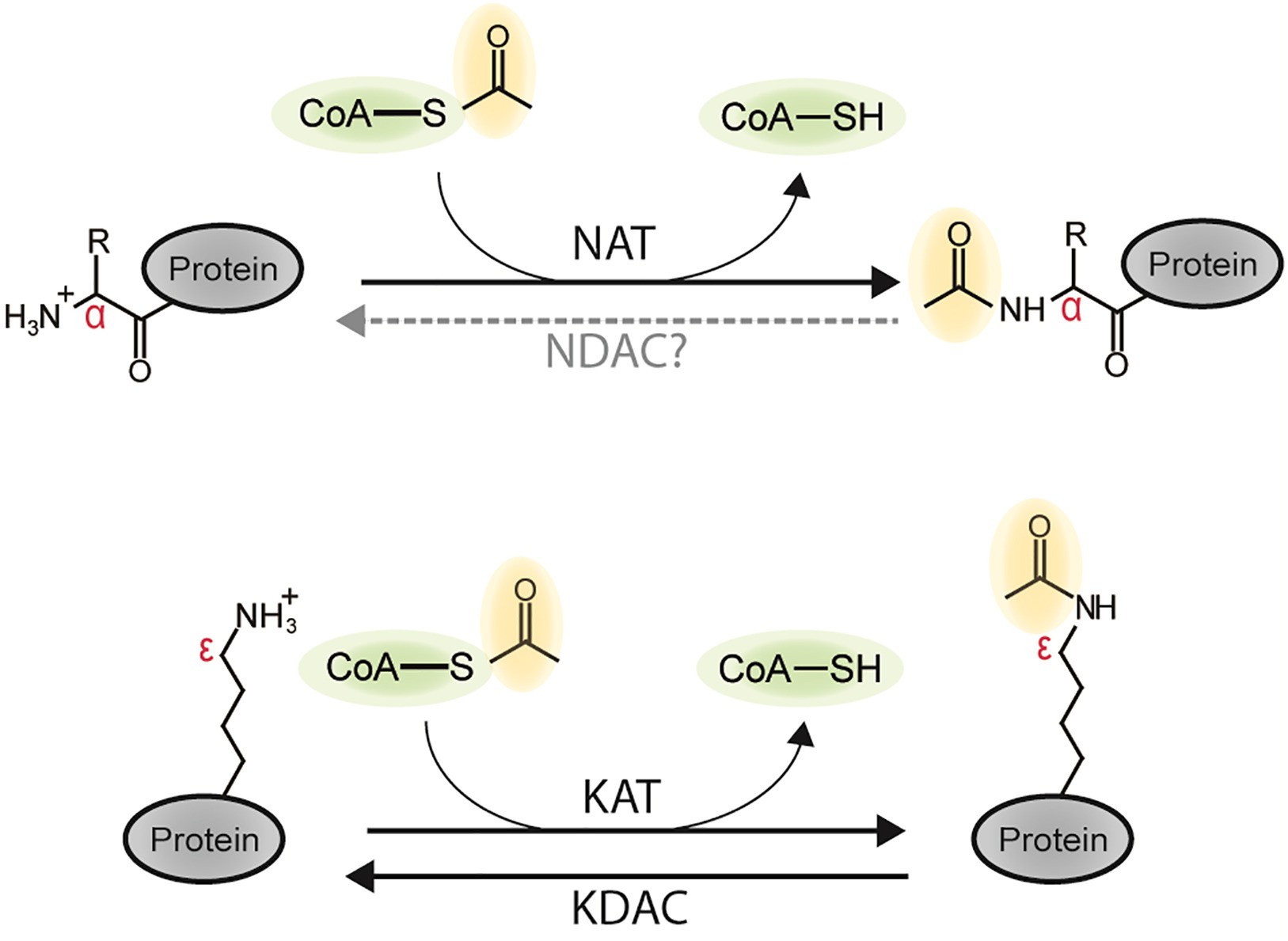
Figure 1. N-terminal and lysine protein acetylation (Aksnes H, et al., 2019).
Why is Proteomics-Acetyl Integrative Analysis Necessary?
Integrative acetyl-proteomics links protein abundance with protein acetylation to reveal how cells regulate function at two levels. Standard proteomics measures how much of each protein exists. Acetylation profiling measures where and how proteins carry a small chemical tag that alters activity, location, or interactions. Many biologically essential changes occur only at the tag level and are invisible to abundance-only assays. Conversely, some protein-level changes occur without modification changes. Joint analysis, therefore, distinguishes regulation by quantity from regulation by modification.
- Integrative analysis reduces false leads. A protein that shows increased acetylation but unchanged abundance points to functional modulation rather than simple upregulation. A protein with both abundance and acetylation changes is more likely to drive a phenotype.
- Integrative studies expose pathway-level coordination. They reveal when coordinated acetylation, expression changes, or both shift entire metabolic or signaling pathways.
- Integrative analysis also improves biomarker and drug-discovery work. It helps identify modification-specific biomarkers that respond to treatment earlier than abundance markers. It clarifies drug mechanisms by showing whether a compound alters protein levels, modification states, or both.
- Integrative workflows increase experimental efficiency. They focus validation on the most promising candidates and reduce wasted resources on changes that lack functional relevance.
Advanced Technologies for Proteomics-Acetyl Integrative Analysis
High-resolution mass spectrometry for deep acetylome profiling
Modern mass spectrometers provide the sensitivity and resolution to detect low-abundance acetylated peptides. Instruments such as Orbitrap and Q-TOF separate and measure peptide ions with high mass accuracy. They resolve peptides that differ by only small mass changes.
DDA/DIA and 4D label-free strategies
- Data-dependent acquisition (DDA) selects the most intense peptides for fragmentation. DDA yields high-quality spectra for identification.
- Data-independent acquisition (DIA) fragments broad windows of ions continuously. DIA provides more complete quantification across samples.
- 4D label-free workflows combine chromatographic time, mass-to-charge, ion mobility, and intensity.
Quantitative approaches: label-based and label-free methods
- Label-based methods use chemical or metabolic tags to compare multiple samples in the same run. Examples include isobaric tags and metabolic labeling. These methods reduce run-to-run variation and allow multiplexing.
- Label-free approaches compare peptide intensities across separate runs. Label-free methods scale well to large cohorts and avoid labeling costs.
Enrichment strategies for acetylated peptides
Acetylated peptides are often low in abundance. Enrichment increases their relative concentration before mass spectrometry. Antibody-based immunoaffinity is the most common approach. High-affinity pan-acetyl antibodies capture acetylated peptides across many proteins. On-bead digestion and elution optimize yield.
Multi-omics integration and systems interpretation
Integrative workflows combine acetylome data with global proteome, phosphoproteome, metabolome, or transcriptome data. Correlation and causality analyses identify co-regulated nodes. Multi-layer networks reveal whether a pathway is regulated by expression, by acetylation, or by both.
Advantages of Our Integrative Analysis Services of Proteomics-Acetyl
- Comprehensive Quantification: Offers both label-based and label-free quantitative platforms.
- High Sensitivity and Specificity: Advanced enrichment methods maximize acetylome coverage.
- Multi-Omics Data Integration: Connects proteomic acetylation patterns with transcriptomic and metabolomic shifts.
- Accurate Bioinformatics Analysis: Uses proprietary pipelines for statistical modeling, pathway mapping, and network visualization.
Workflow for Integrative Analysis Services of Proteomics-Acetyl
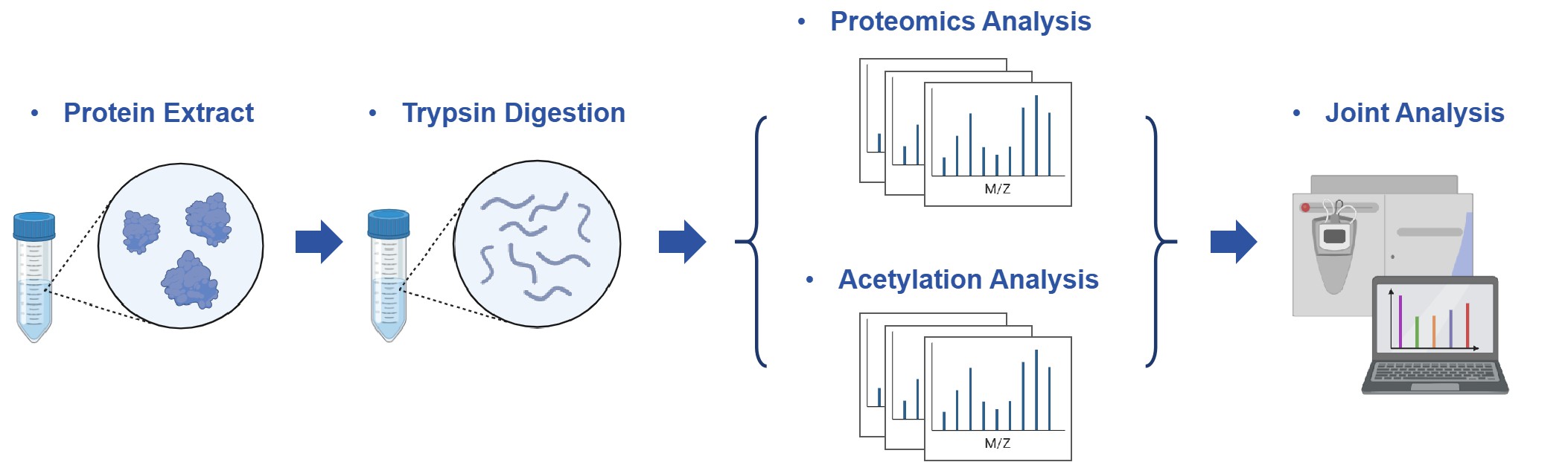
Deliverables and Reporting Standards
Data Deliverables
- Raw MS Data: High-resolution mass spectrometry files in the original instrument format.
- Processed Quantitative Data: Tables showing identified acetylation sites, peptide sequences, protein IDs, intensity values, and statistical significance.
- Integrated Multi-Omics Results: Combined insights from acetyl-proteomics with other omics layers, such as transcriptomics or metabolomics.
Reporting Standards
- Comprehensive Analysis Report: Includes methodology, experimental workflow, quality control results, and key findings.
- Data Visualization: Publication-ready charts, heatmaps, volcano plots, and pathway enrichment maps to simplify complex datasets.
- Pathway and Functional Insights: Biological interpretation of acetylation patterns and their roles in cellular regulation or disease mechanisms.
Applications of Integrated Proteomics-Acetyl Analysis
- Epigenetic Regulation Studies: Quantify histone acetylation and its impact on chromatin accessibility.
- Signal Transduction Mapping: Characterize acetylation-mediated activation of key signaling pathways.
- Metabolic Network Reconstruction: Link acetylation dynamics to central carbon metabolism and energy balance.
- Biomarker Discovery: Identify novel acetylation-driven candidates for translational research.
- Drug Target Validation: Evaluate how acetylation patterns respond to therapeutic interventions.
Simple Requirements
| Category | Requirement |
| Sample Types | Cell lysates, tissue extracts, serum, plasma, or other biological samples. |
| Sample Amount | Recommended ≥ 200 µg total protein per sample. |
| Protein Concentration | At least 1 µg/µL for optimal enrichment and detection. |
| Sample Purity | Avoid contamination from detergents, salts, or nucleic acids. |
| Storage Conditions | Store at -80°C to maintain protein integrity and prevent acetylation loss. |
| Shipping | Use dry ice or other low-temperature methods to ensure sample stability. |
Why Choose Creative Proteomics
- Expert team. Our scientists have more than 20 years of experience in proteomics and post-translational modification analysis.
- Comprehensive platforms. We perform tagging-based quantification (TMT and SILAC) and label-free workflows on modern high-resolution mass spectrometers.
- Customized experimental design. Our staff tailor sample preparation, enrichment, and quantification strategies to each project goal.
- Rigorous quality control. We include technical replicates, spike-in controls, and standardized metrics to ensure data integrity.
- Robust data analysis. We provide site-level quantification, differential testing, pathway enrichment, and interaction networks in publication-ready form.
FAQ
-
Q1: Which categories of proteins are most frequently acetylated, and what pathways are impacted?
A1: Acetylation occurs not only on histones but also on transcription factors, metabolic enzymes, chaperones, and cytoskeletal proteins. It regulates mitochondrial metabolism, cell-cycle progression, gene expression, signal signaling pathways, and immune responses.
-
Q2: What normalization methods are commonly applied to acetyl-proteome data?
A2: Normalization uses total-ion current, median centering, or variance stabilizing transformations. Label-based experiments employ isotopic ratio normalization. Choice depends on experimental design.
-
Q3: How is cross-talk between acetylation and other PTMs assessed?
A3: Cross-talk assessment requires parallel profiling of other PTMs and integrative network analysis. Temporal sampling enhances detection of sequential modification events.
Demo
Demo: Quantitative Analysis of Global Proteome and Lysine Acetylome Reveal the Differential Impacts of VPA and SAHA on HL60 Cells
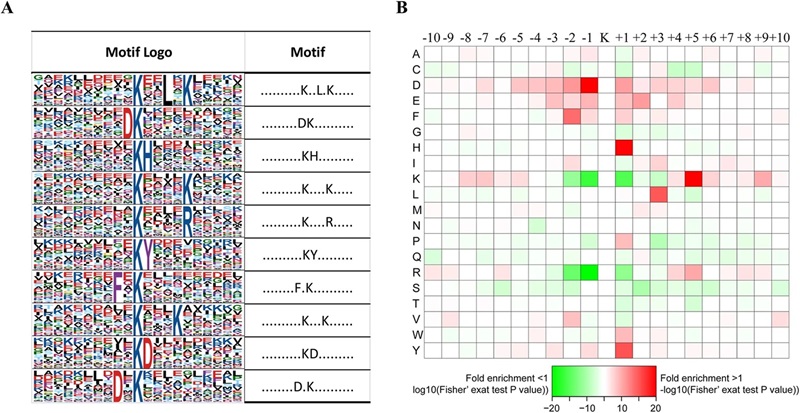
Figure 2. Properties of all the identified Kac peptides (Zhu X Y, et al., 2016).
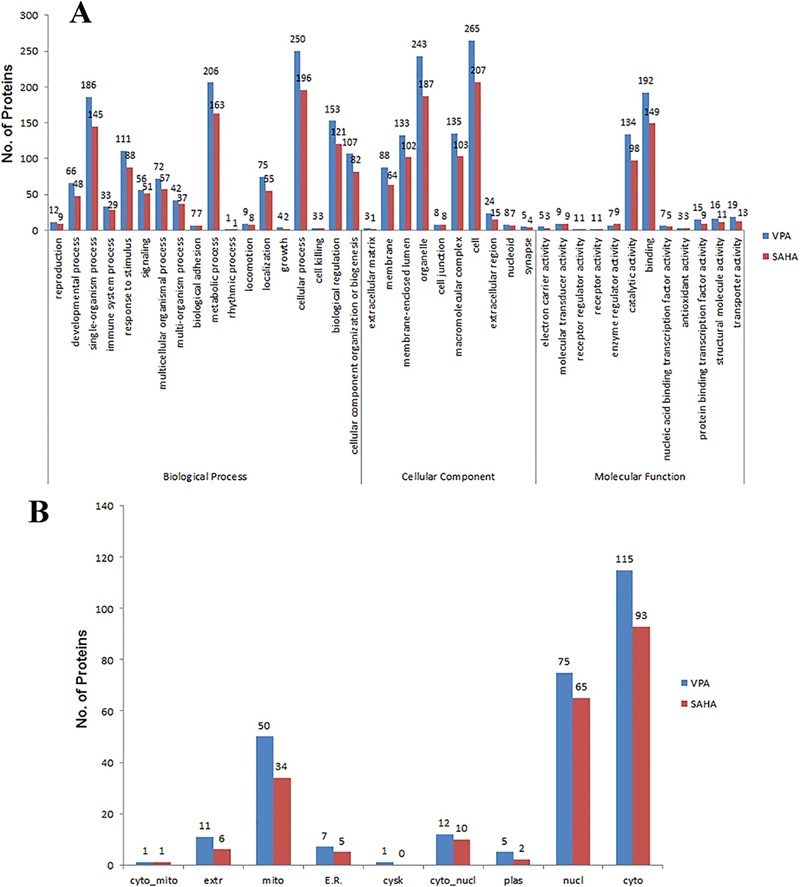
Figure 3. Gene ontology (GO) classification analysis (Zhu X Y, et al., 2016).
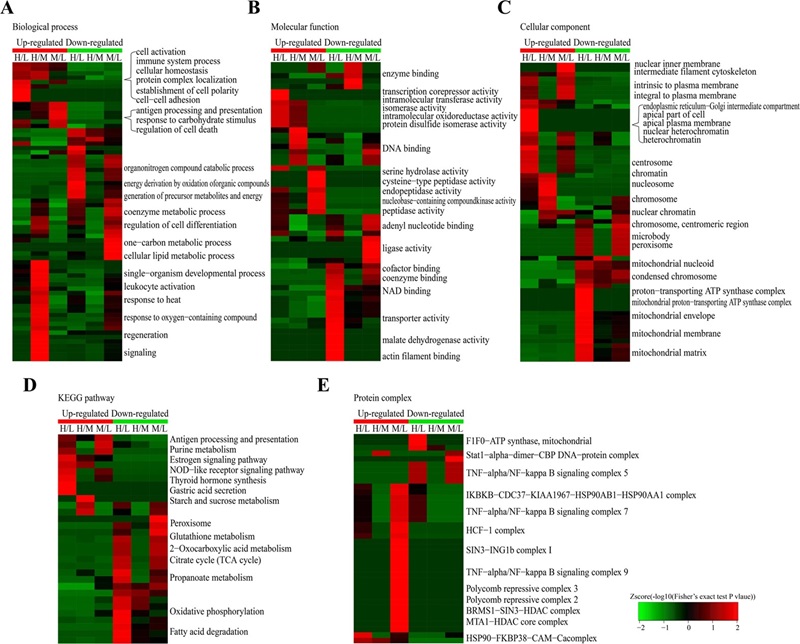
Figure 4. Enrichment and clustering analysis of the quantitative acetylome data sets in VPA and SAHA treated AML HL60 cells based on biological process (Zhu X Y, et al., 2016).
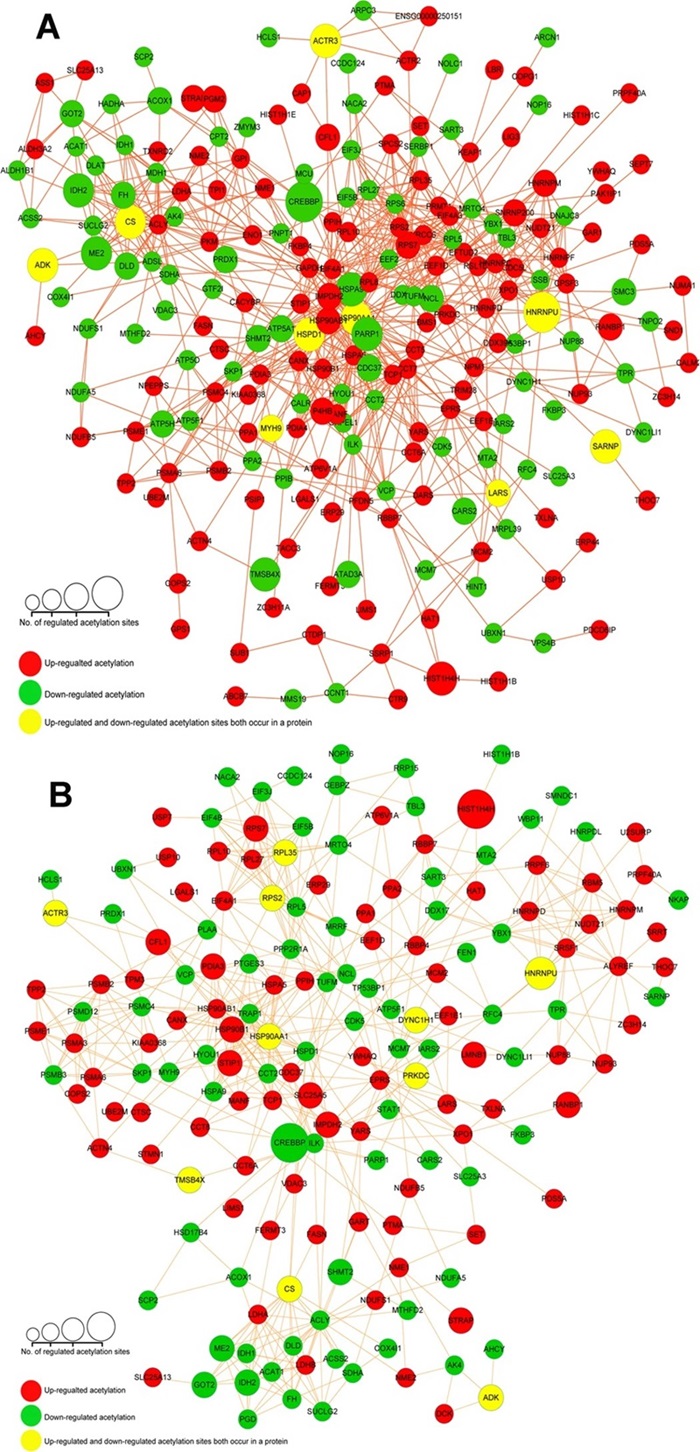
Figure 5. Protein-protein interaction network analyses for the different expressed 268 acetylated proteins upon VPA treated (A) and SAHA treated (B) in AML HL60 cells (Zhu X Y, et al., 2016).
-
Case Study
Case: High-intensity interval training remodels the proteome and acetylome of human skeletal muscle.
Abstract:
Exercise elicits systemic metabolic benefits. Skeletal-muscle adaptation to exercise is mediated by changes in protein abundance and by post-translational modifications. Lysine acetylation is a widespread PTM that modulates mitochondrial function, chromatin state, and metabolic enzymes. The study asked how a defined programme of high-intensity interval training (HIIT) reshapes both the global proteome and the lysine acetylome of human skeletal muscle to reveal mechanistic links between training and metabolic adaptation.
Methods
- Human cohort and intervention: Healthy adult volunteers completed a supervised HIIT intervention with pre- and post-training skeletal muscle biopsies (paired design). The experimental design enabled within-subject comparison of proteome and acetylome changes induced by exercise training.
- Proteomics and acetylomics workflow: Muscle samples were processed for protein extraction and tryptic digestion. Global proteome measurements were acquired by high-resolution LC-MS/MS. Lysine-acetylated peptides were enriched (anti-acetyl-lysine immunoaffinity) prior to MS to map site-specific acetylation. Quantitative pipelines were applied to detect differential protein abundance and differential acetylation at the site level.
- Data analysis: Identified proteins and acetyl-sites were subjected to statistical testing, gene ontology and pathway enrichment, and mitochondrial/complex-level annotation to interpret functional trends. Datasets were deposited in public repositories for reuse.
Results
- Proteome coverage: The authors quantified ~3,168 proteins in human skeletal muscle and identified exercise-responsive changes across metabolic and contractile systems.
- Acetylome coverage: They detected ~1,263 lysine acetylation sites on ~464 acetylated proteins. The acetylome exhibited distinct training-associated modulation.
- Mitochondrial acetylation: HIIT increased acetylation on numerous mitochondrial proteins, with an enrichment in proteins of complex V (ATP synthase), suggesting altered regulation of oxidative phosphorylation via acetylation.
- Contractile and calcium-handling proteins: Proteomic changes implicated excitation–contraction coupling and altered myofibrillar calcium sensitivity after training. These changes align with functional adaptations in muscle contractility.
- Histone acetylation: Select histone acetyl-sites were exercise-responsive, pointing to potential transcriptional remodeling associated with training.
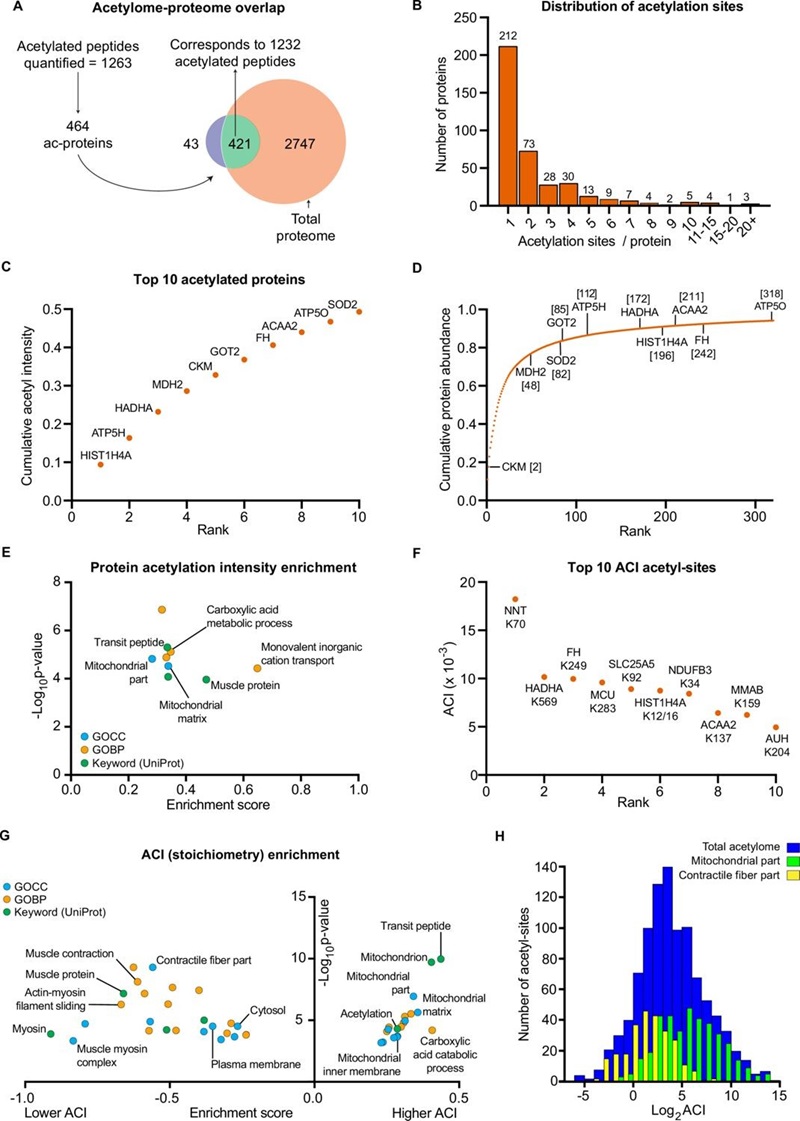
Figure 6. The human skeletal muscle acetylome displays higher stoichiometry on mitochondrial proteins and lower stoichiometry on contractile proteins.
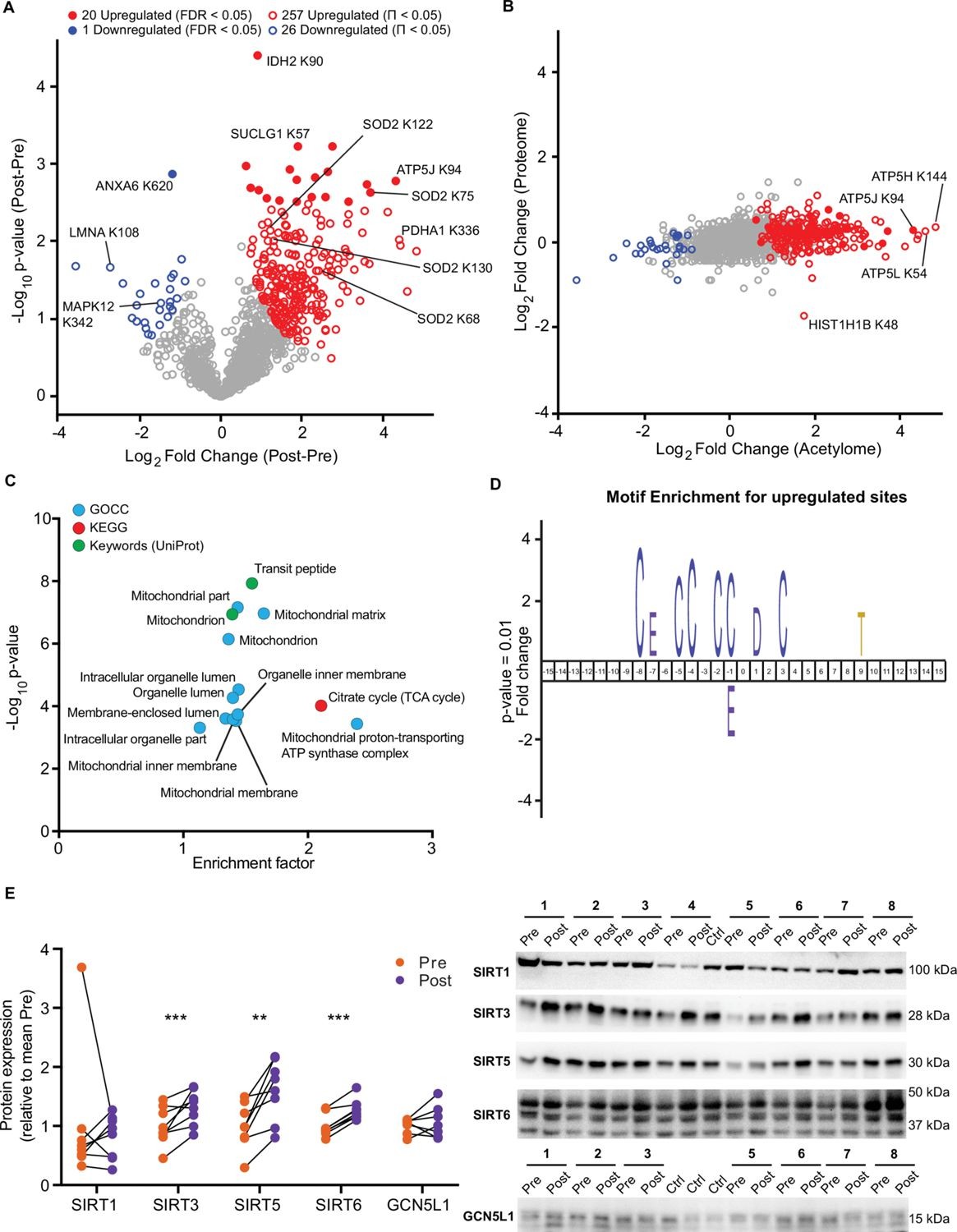
Figure 7. HIIT increases acetylation of mitochondrial and TCA cycle proteins concomitantly with an increase in SIRT3 abundance.
Conclusion
The study demonstrates that short-term HIIT drives both global changes in protein abundance and site-specific changes in lysine acetylation in human skeletal muscle. The data support a model in which training-induced metabolic and contractile adaptations are mediated partly through reversible acetylation of mitochondrial and nuclear proteins. The integrated proteome + acetylome resource constitutes a hypothesis-generating atlas for future mechanistic studies on exercise, metabolism, and health.
Related Services
References
- Aksnes H, Ree R, Arnesen T. Co-translational, post-translational, and non-catalytic roles of N-terminal acetyltransferases. Molecular cell, 2019, 73(6): 1097-1114.
- Popova L, Carr R A, Carabetta V J. Recent Contributions of Proteomics to Our Understanding of Reversible Nε-Lysine Acylation in Bacteria. Journal of proteome research, 2024, 23(8): 2733-2749.
- Shang S, Liu J, Hua F. Protein acylation: mechanisms, biological functions and therapeutic targets. Signal transduction and targeted therapy, 2022, 7(1): 396.

