Autoantigen Microarray Profiling Service
What Is the Autoantigen Microarray?
An autoantigen microarray is an arrayed platform that presents multiple antigenic entities for interrogation by patient immunoglobulins. The antigenic entities may be recombinant proteins, synthetic peptides, phage-displayed peptides, or fractions of native proteomes. The arrayed features serve as baits. Serum or plasma antibodies bind cognate epitopes. Detection occurs via labeled secondary reagents and high-resolution imaging. Data are captured as intensity matrices. Subsequent informatics pipelines transform intensities into biomarker candidates.
There are three principal formats in routine use. The first is the recombinant/peptide array. Known antigens are printed in defined locations. This format facilitates rapid translation to targeted assays. The second is the native fraction array. Native proteins or chromatographic fractions are spotted to preserve post-translational modifications (PTMs) and conformational epitopes. The third is the reverse-capture (antibody) array. Monoclonal capture antibodies immobilize native antigens from lysates. The captured antigens are probed with patient IgG, and identities are intrinsic to the capture probes. Each format supplies distinct advantages for discovery or validation.
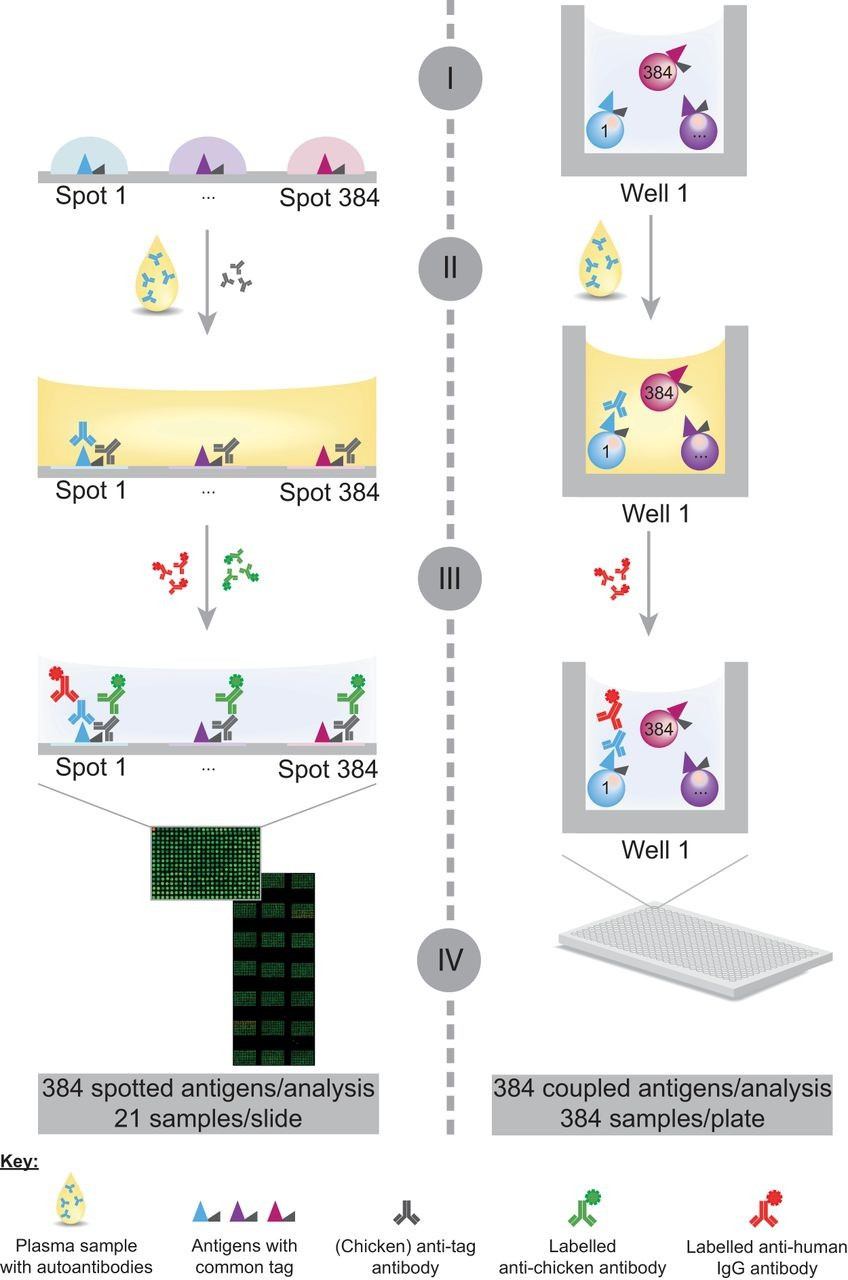
Figure 1. Schematic representation of the assay workflow on planar and bead-based antigen arrays for profiling autoantibody responses (Ayoglu B, et al. 2013).
Why Choose Autoantibody Profiling for Biomarker Discovery?
Antibodies function as endogenous signal amplifiers. A minute, tissue-restricted antigen can elicit a sustained humoral response. The humoral response persists longer than many free antigens in circulation. Therefore, autoantibody signatures often provide earlier or more robust detection than direct antigen assays. Multiplexed arrays increase diagnostic sensitivity by aggregating responses to multiple antigens. Autoantibody profiles can stratify disease subtypes. They can reveal pathophysiologic mechanisms, such as neo-epitope formation or PTM-dependent autoimmunity. Finally, microarray formats permit high-throughput screening with low sample consumption.
Our Autoantigen Microarray Profiling Services
Creative Proteomics provides end-to-end autoantigen microarray solutions. We offer discovery, focused validation, and assay translation services. Our platform portfolio includes recombinant/peptide arrays, phage-display derived arrays, native fraction arrays, and reverse-capture antibody arrays. We supply turnkey project design, sample QC, array hybridization, imaging, bioinformatics, and report generation. We also support orthogonal validation by mass spectrometry, co-immunoprecipitation, and multiplex ELISA development. Project scopes are customizable.
Key service attributes:
- Multi-format platform selection to match scientific aims.
- Low-volume assays compatible with serum/plasma.
- Integrated bioinformatics with reproducible code and cross-validation.
- Translation pathways to multiplex ELISA and bead-based assays.
Workflow for Autoantigen Microarray Profiling Services
- Sample preparation. Samples are inspected, aliquoted, and evaluated for hemolysis, lipemia, and integrity.
- Array selection and preparation. The optimal platform is chosen and configured. Antigens are printed or captured according to the selected format.
- Hybridization and detection. Patient immunoglobulins are incubated with arrays. Secondary detection reagents produce a measurable signal.
- Imaging and raw data capture. High-resolution scanners generate raw intensity files.
- Preprocessing and normalization. Background correction, dye-bias adjustment, and probe QC are applied.
- Statistical analysis and modelling. Differential reactivity tests, false discovery rate control, and classifier construction are performed.
- Orthogonal validation (optional). Mass spectrometry, Co-IP, or ELISA validate candidate antigens..
- Assay conversion (optional). Select candidates are ported to multiplex ELISA or bead arrays for analytical validation.
- Reporting and delivery. Visualizations, tables, and interpretive text are delivered with raw and processed data.
Deliverables and Reporting Standards
- Raw data files. Unprocessed scanner outputs in vendor format.
- Processed matrices. Background-corrected and normalized intensity tables in CSV format.
- Quality control reports. Per-probe and per-array QC metrics, including replicate CVs and control probe performance.
- Statistical summaries. Differential reactivity tables with fold change, p-value, and adjusted p-value.
- Visualizations. Heatmaps, volcano plots, hierarchical clustering, and ROC curves.
- Candidate marker lists. Ranked panels with selection rationale and annotation.
- Methodological documentation. Detailed protocols, reagent lists, and batch metadata.
Applications of Autoantigen Microarray Profiling
- Oncology: Autoantibody signatures can reveal tumor-associated antigens produced during cancer development. These profiles help researchers discover potential biomarkers for early cancer detection, track disease progression, and monitor treatment responses.
- Autoimmune Diseases: The immune system mistakenly attacks the body's proteins in autoimmune disorders. Autoantigen microarrays enable researchers to map autoantibody profiles linked to specific diseases, such as systemic lupus erythematosus (SLE), rheumatoid arthritis, and multiple sclerosis.
- Infectious Diseases: Viral and bacterial infections often trigger immune responses that produce cross-reactive autoantibodies. Autoantigen microarrays are widely used to study immune system changes after infections such as COVID-19, hepatitis, or influenza.
- Drug Development and Biotherapeutic Safety: In drug discovery, monitoring the body's immune response is critical, especially when developing biologic drugs such as monoclonal antibodies.
- Pharmacodynamics and Treatment Monitoring: Changes in autoantibody levels over time can indicate whether a treatment is effective. By profiling immune responses before and after therapy, researchers can evaluate treatment efficacy, track patient recovery, and identify early signs of relapse or resistance.
- Toxicology and Safety Assessment: Certain drugs or environmental exposures can unintentionally trigger immune-related side effects. Autoantigen microarrays provide a high-throughput solution for screening immune toxicity risks, identifying potential safety concerns earlier in the development pipeline, and ensuring regulatory compliance.
Simple Requirements
| Accepted Sample Types | Serum, plasma (EDTA or heparin), cerebrospinal fluid (CSF), saliva, or purified IgG. |
| Minimum Volume | 10–50 µL per sample. |
| Storage Conditions | Store at −80 °C. Avoid more than two freeze-thaw cycles. |
| Sample Quality | Use clear, non-hemolyzed, non-lipemic samples. Document collection method and time. |
Why Choose Creative Proteomics
- Cutting-edge platform: High-throughput autoantigen microarray technology with industry-leading sensitivity.
- Comprehensive services: Covering multiple disease models and research areas, from discovery to validation.
- Expert scientific team: Rich experience in proteomics and immunology research.
- Client-focused solutions: Customized experimental design and data interpretation tailored to your research goals.
FAQ
-
Q1: How does an autoantigen microarray differ from ELISA?
A1: Both assays detect antibody binding. ELISA measures one antigen per reaction and is inherently single-plex. Microarrays measure many antigens in one experiment and thus deliver multiplexed profiles that improve discovery throughput and enable serological signatures that are impossible with single-analyte tests. Reviews show microarrays can markedly increase diagnostic information relative to single-target ELISA.
-
Q2: How are autoantigen microarrays used in systems biology and multi-omics studies?
A2: Autoantigen microarrays can be integrated with proteomics, transcriptomics, and metabolomics to reveal immune-related molecular mechanisms. By combining autoantibody profiling with mass spectrometry-based proteomics, researchers can correlate immune signatures with protein abundance, PTMs, and pathway dysregulation. Multi-omics integration enhances biomarker discovery and provides a more comprehensive understanding of disease pathogenesis.
-
Q3: How does the antigen source affect experimental outcomes?
A3: Antigen origin—recombinant, synthetic peptide, or native protein—directly affects antibody binding specificity:
Recombinant antigens enable broad screening, but may lack PTMs.
Synthetic peptides allow epitope mapping but lose conformational structure.
Native proteins preserve structural integrity and PTMs, increasing detection accuracy for clinically relevant epitopes.
-
Q4: What public databases or resources are available for self antigen microarray data?
A4: ImmPort: NIH repository for immune profiling datasets
ArrayExpress: Gene Expression Omnibus (GEO) microarray datasets
HuProt Database: Comprehensive reference for human protein arrays
Demo
Demo: SARS-CoV-2 multi-antigen protein microarray for detailed characterization of antibody responses in COVID-19 patients
Developed a multi-antigen protein microarray containing spike, nucleocapsid and variant domains to compare antibody repertoires across acute and convalescent COVID-19 patients and to resolve variant-specific reactivity. The study shows microarray concordance with ELISA and highlights variant epitope detection.
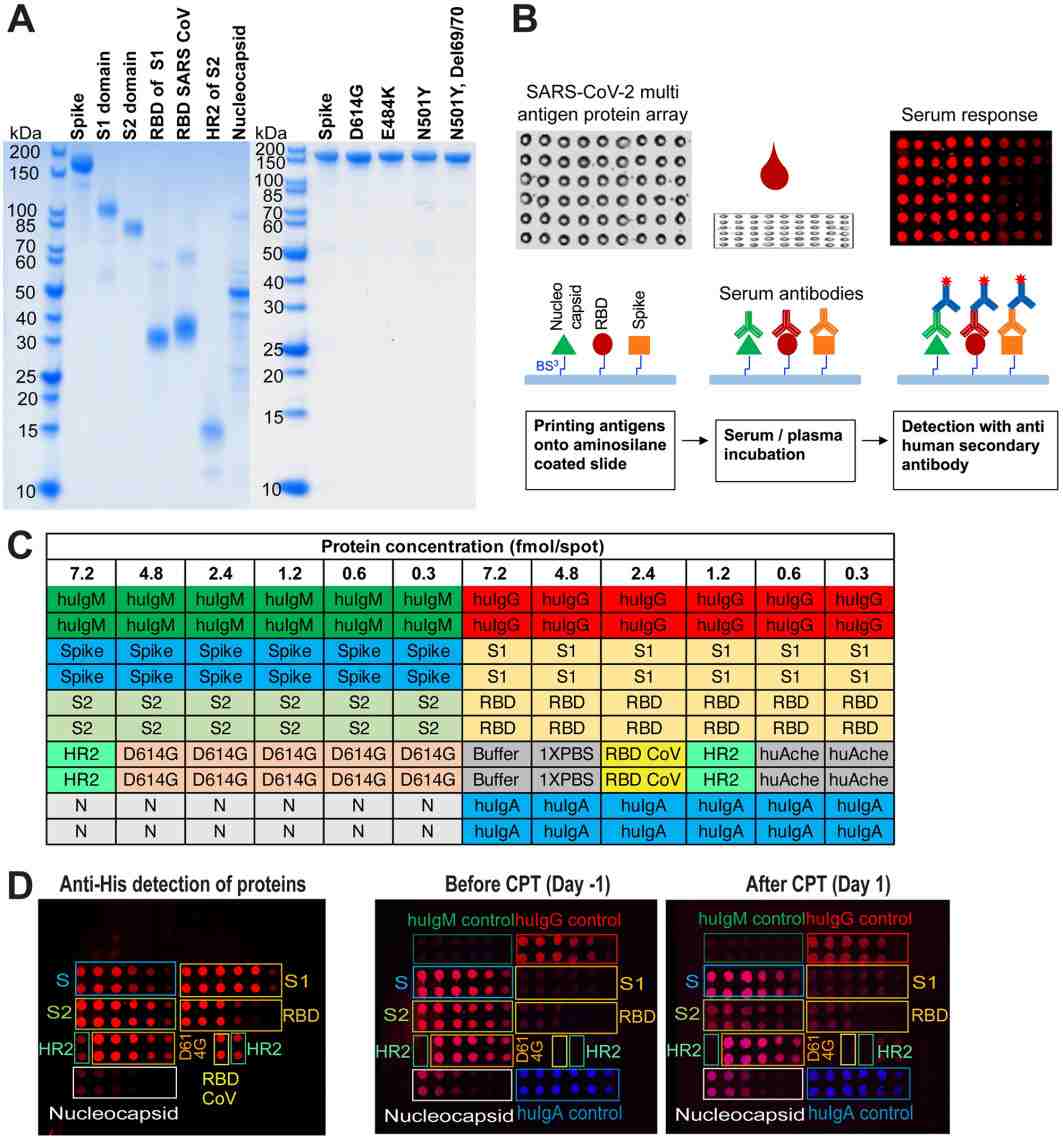
Figure 2. SARS-CoV-2 multi-antigen protein array production and processing (Celikgil A, et al., 2023).
-
Case Study
Case: High-Throughput Antigen Microarray Identifies Longitudinal Prognostic Autoantibody for Chemoimmunotherapy in Advanced Non-Small Cell Lung Cancer
Abstract:
Immune checkpoint inhibitors (ICIs), often combined with chemotherapy (chemoimmunotherapy), are standard treatments for advanced non-small cell lung cancer (aNSCLC). However, heterogeneous responses and acquired resistance remain major clinical problems. Autoantibodies in plasma may be circulating biomarkers predicting response or progression during ICI-based therapy. The study leveraged high-density antigen microarrays to search for longitudinally dynamic autoantibody (AAb) signatures associated with treatment response and progression in aNSCLC. To discover, verify, and validate autoantibody biomarkers that (1) associate with early treatment response and (2) predict progression-free survival (PFS) in aNSCLC patients receiving chemoimmunotherapy. The investigators aimed to identify pre-treatment and on-treatment AAb panels useful for prognostic monitoring and potential clinical translation.
Methods
- Study design: Three-phase workflow — discovery, verification, and validation — using paired longitudinal plasma samples collected before and during therapy.
- Samples: 528 plasma samples from 267 aNSCLC patients (paired time points) were analyzed overall; additional FFPE tumor specimens and public transcriptomic datasets were used for orthogonal validation.
- Discovery platform: A HuProt high-density protein microarray (~21,000 human proteins) was used to screen for candidate prognostic AAbs in the discovery phase.
- Verification & validation: A focused aNSCLC microarray (507 candidate AAbs) and subsequent ELISA verification (11 → final candidates) were used across independent sample sets and time points (T0 baseline, T1 on-treatment, T2 progression where available). Statistical pipelines included differential analysis (limma) and univariate Cox regression for PFS. Orthogonal validation included immunohistochemistry (IHC) on FFPE and interrogation of public ICI datasets (GEO).
Results
- Candidate identification: From the HuProt discovery screen and stepwise verification, the authors narrowed candidates to a small set of prognostic AAbs.
- Validated markers: The study identified and validated two pre-treatment AAbs (MAX and DHX29) and two on-treatment predictive AAbs (MAX and TAPBP) correlated with early responses and PFS in chemoimmunotherapy patients.
- Dynamics and predictive performance: MAX autoantibody levels increased in responders during treatment and followed distinct kinetics in non-responders; MAX protein and mRNA levels (IHC / transcriptomic datasets) also discriminated PFS groups (statistically significant, p < 0.05). ROC and Cox analyses in verification/validation cohorts supported the prognostic association of these AAbs.
- Orthogonal validation: IHC on tumor FFPE and GEO immunotherapy transcriptomic datasets analyses corroborated some AAb targets' clinical relevance (MAX, etc.).
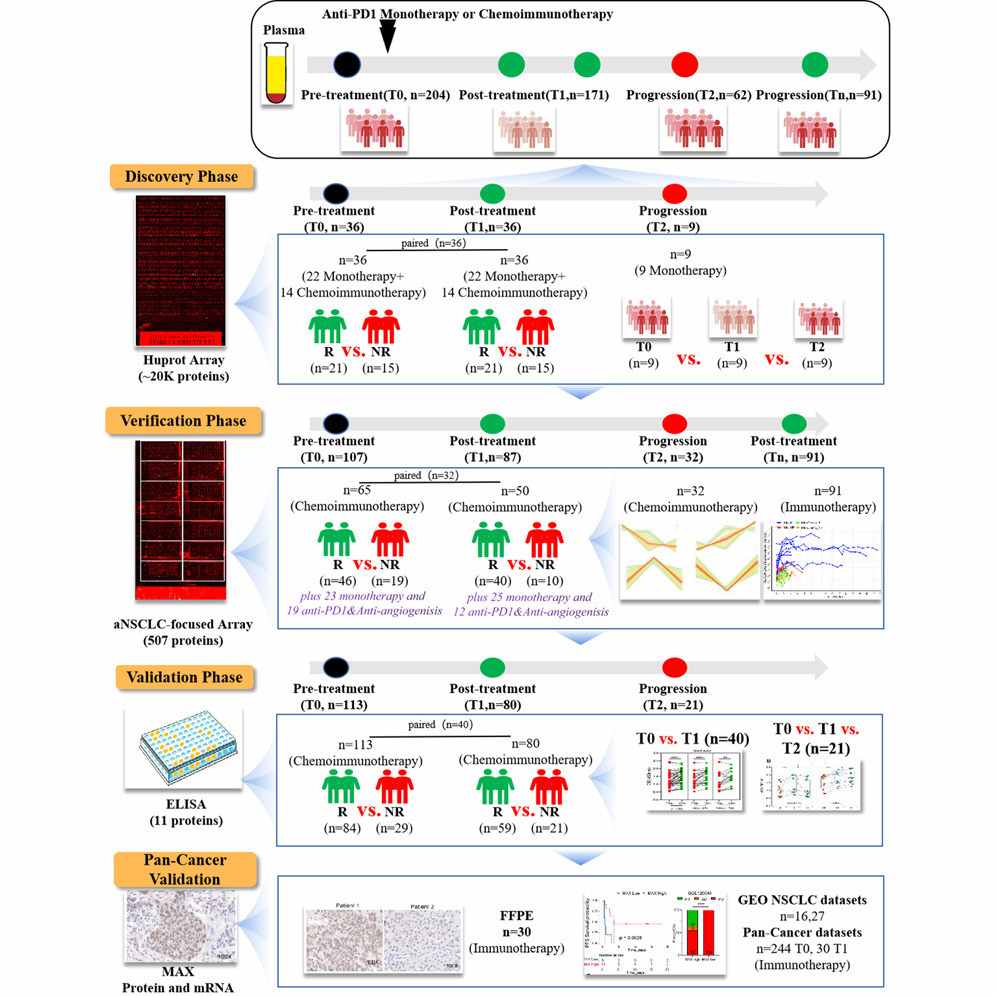
Figure 3. Graphical abstract for high-throughput antigen microarray identifies longitudinal prognostic autoantibody for chemoimmunotherapy.
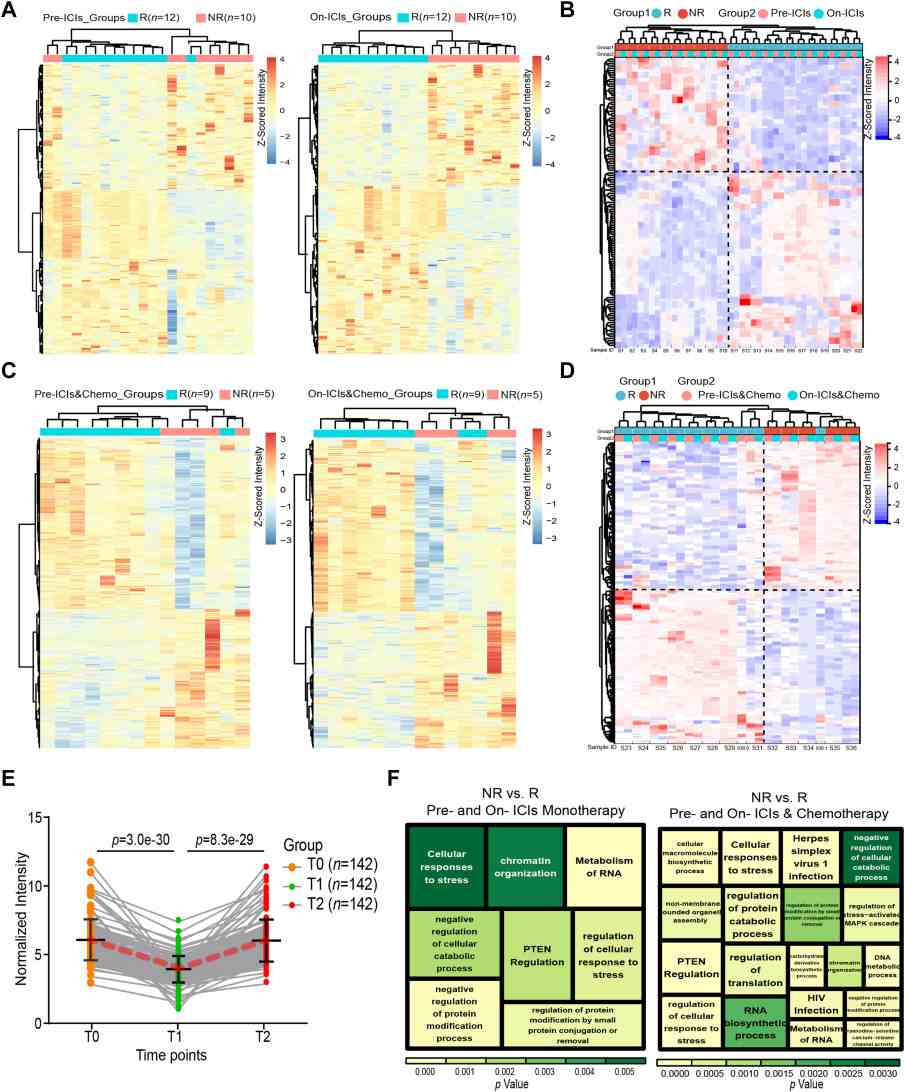
Figure 4. High-density microarrays for identifying differential AAbs (p < 0.05) in the discovery phase (n = 36).
Conclusion
The authors conclude that high-density antigen microarray profiling can identify longitudinal autoantibody biomarkers that predict response and progression in aNSCLC patients treated with chemoimmunotherapy. Specific AAbs emerged as candidate prognostic / monitoring markers. The study demonstrates a scalable discovery, verification to validation pipeline that combines proteome-wide screening with focused arrays, ELISA confirmation, and orthogonal tissue/transcriptome validation — supporting further prospective evaluation and translational development.
Related Services
References
- Celikgil A, et al. SARS-CoV-2 multi-antigen protein microarray for detailed characterization of antibody responses in COVID-19 patients. PLoS One, 2023, 18(2): e0276829.
- Kuzumi A, et al. Comprehensive autoantibody profiling in systemic autoimmunity by a highly-sensitive multiplex protein array. Frontiers in Immunology, 2023, 14: 1255540.
- Dai L, et al. High-Throughput antigen microarray identifies longitudinal prognostic autoantibody for chemoimmunotherapy in advanced Non-Small cell lung Cancer. Molecular & Cellular Proteomics, 2024, 23(5): 100749.
- Ayoglu B, et al. Autoantibody profiling in multiple sclerosis using arrays of human protein fragments. Molecular & Cellular Proteomics, 2013, 12(9): 2657-2672.

