Aptamer-Based Proteomics Profiling Service
What is Aptamer-Based Proteomics Profiling?
Aptamer-based proteomics profiling is a molecular detection and quantification technique that utilizes synthetic oligonucleotides, known as aptamers, to bind protein targets selectively. These aptamers are generated through Systematic Evolution of Ligands by Exponential Enrichment (SELEX), which allows the selection of molecules with high specificity and affinity toward target proteins. Unlike antibodies, aptamers are chemically synthesized, reducing batch-to-batch variability and ensuring consistent performance. The technology enables researchers to monitor hundreds to thousands of proteins in a single assay, providing a global view of the proteome.
High-plex aptamer-based proteomics allows simultaneous measurement of thousands of proteins in a single experiment. The technology preserves native protein conformations and post-translational modifications, providing biologically meaningful data. It is particularly effective for membrane proteins, low-abundance targets, and cell-surface markers that are challenging for conventional methods.
Why Choose Aptamers for Proteomics?
Aptamers possess several properties that make them particularly suitable for proteomic analysis:
- High specificity and affinity: Comparable to or exceeding monoclonal antibodies.
- Thermal and chemical stability: Aptamers can withstand harsh conditions, making them robust for diverse experimental setups.
- Reduced immunogenicity: Unlike antibodies, aptamers do not provoke immune responses, enabling their use in clinical research.
- Batch reproducibility: Chemical synthesis ensures consistent quality and reduces variability in large-scale studies.
- Rapid customization: Aptamers can be generated against challenging targets, including non-immunogenic or toxic proteins.
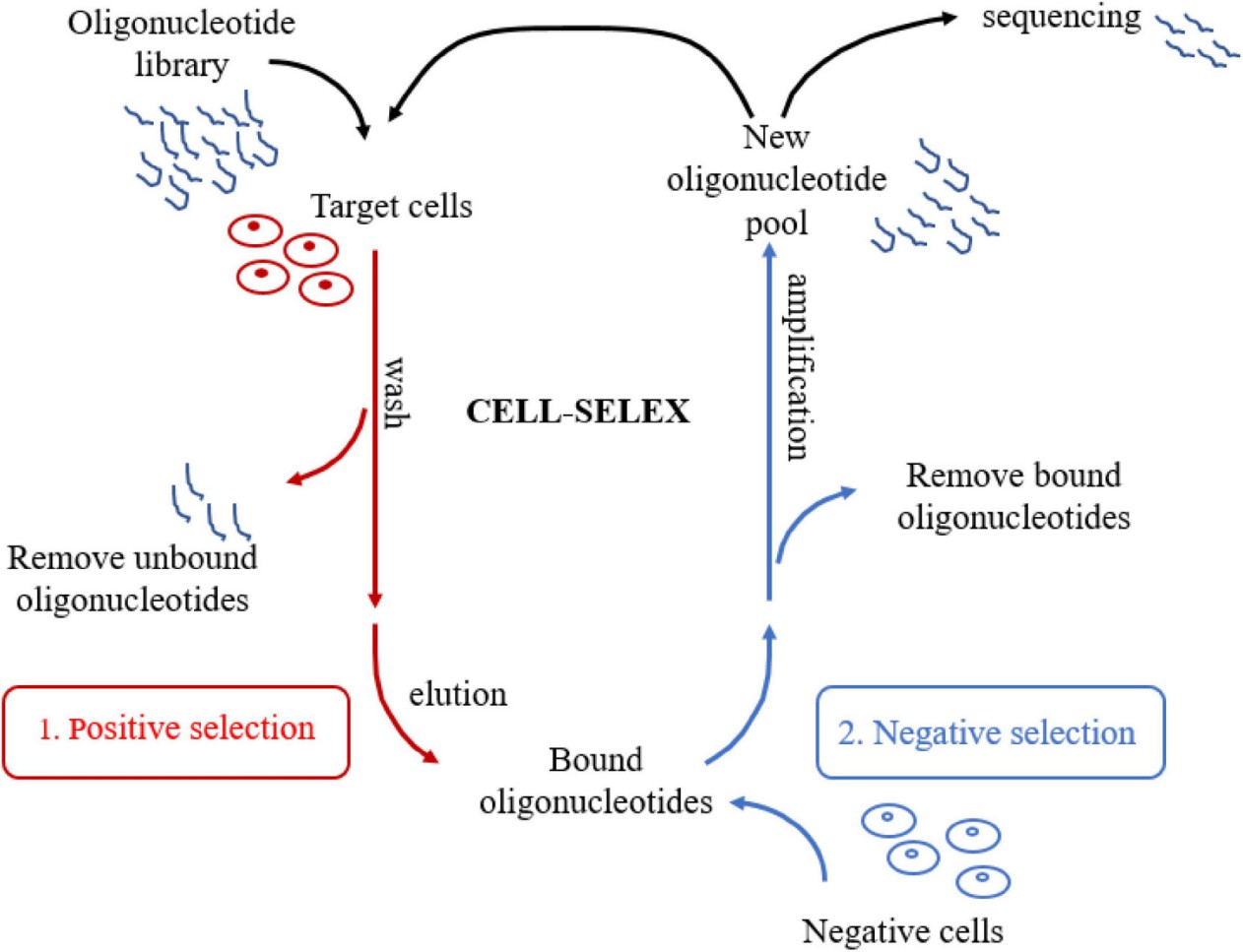
Figure 1. Schematic representation of CELL-SELEX (Huang J, et al., 2021).
Key Technology for Aptamer-Based Proteomics Profiling
SELEX
SELEX provides the fundamental selection process. The laboratory performs standard SELEX for purified targets. The team employs Cell-SELEX for whole-cell targets. The group applies in vivo SELEX for tissue-directed selection when applicable. The platform includes microfluidic and automated SELEX variants for throughput and reproducibility. The laboratory implements counter-selection to remove non-specific binders. The scientists monitor enrichment across iterative rounds.
Next-generation sequencing
NGS quantifies sequence enrichment. Bioinformatics identifies candidate aptamer families. Motif analysis detects conserved structural elements. The computational pipeline estimates relative abundance and convergence. The team annotates potential off-target motifs. The data supports rational truncation and affinity maturation.
Target identification by mass spectrometry
Affinity pull-downs with candidate aptamers precede LC-MS/MS analysis. The proteomic workflow employs high-resolution instruments. Tandem mass spectrometry achieves peptide sequencing and protein identification. Target deconvolution uses label-free quantification or isobaric labeling when needed. The approach enables identification of post-translational modifications and interacting partners.
Assay development and orthogonal validation
The group develops orthogonal assays for confirmation. Flow cytometry, immunofluorescence, and targeted PRM validate localization and abundance. Functional assays examine biological relevance. The integrated pipeline ensures that candidate aptamers translate into usable reagents.
Compare Aptamer-Based Profiling with Mass Spectrometry Proteomics
| Feature | Aptamer-Based Profiling | Mass Spectrometry Proteomics |
| Principle | Uses short, single-stranded DNA or RNA molecules that bind targets with high specificity. | Measures protein mass and fragments to identify and quantify proteins directly. |
| Target Knowledge Required | Can discover unknown proteins; works with cell surfaces and complex samples. | Hypothesis-free; no prior knowledge needed. |
| Sensitivity | High affinity for low-abundance proteins; can detect rare targets on cell surfaces. | High dynamic range but may miss extremely low-abundance proteins without enrichment. |
| Specificity | Can distinguish closely related isoforms and conformations; chemical modifications improve selectivity. | Very high specificity at peptide level; can detect sequence variants and modifications. |
| Throughput | High; multiple aptamers can be screened simultaneously. | High, but dependent on instrument time and sample preparation. |
| Post-Translational Modifications (PTMs) | Indirect detection possible if aptamer binds modified sites, but not comprehensive. | Excellent; can identify phosphorylation, acetylation, glycosylation, and other modifications. |
| Ease of Optimization | Aptamers can be chemically modified and iteratively optimized in vitro. | Optimization relies on instrument settings and sample prep; no reagent modification possible. |
| Sample Flexibility | Works with purified proteins, cell surfaces, tissues, and complex mixtures. | Works with diverse samples but may require enrichment for low-abundance targets. |
| Ideal Use Cases | Biomarker discovery, receptor mapping, early-stage diagnostics, cell-type specific targets. | Comprehensive proteome mapping, PTM analysis, interactome studies. |
Workflow for Our Aptamer-Based Proteomics Profiling Service
- Sample preparation: A laboratory technologist inspects sample integrity. The group performs basic QC assays. The team documents concentration, viability for cells, and storage history.
- Library preparation and SELEX: A scientist synthesizes a randomized nucleic acid library. The platform initiates iterative selection cycles. The laboratory conducts positive and negative selection steps. The group monitors enrichment by qPCR or pilot sequencing.
- High-throughput sequencing: A bioinformatician processes NGS reads. The pipeline ranks candidate families by enrichment and diversity. The team selects representative sequences for synthesis.
- Binding characterization: An analyst measures affinity and kinetics for each candidate. The laboratory performs specificity panels against control cell lines or proteins. The team trims and optimizes sequences for stability and performance.
- Affinity pull-down and mass spectrometry: A chemist immobilizes top candidates for pull-downs. The proteomics unit performs LC-MS/MS analysis. The group identifies bound proteins and quantifies enrichment versus controls.
- Reporting and delivery: A project lead compiles a technical dossier. The dossier contains sequence files, NGS metrics, MS identifications, binding curves, and validation data. The group delivers raw data upon request.

Applications of Aptamer-Based Proteomics Profiling
- Biomarker discovery: The platform identifies cell-type specific surface markers. The method aids discovery in oncology, immunology, and neurology. The workflow supports subsequent biomarker qualification.
- Target validation for therapeutics: Aptamers assist in receptor mapping and target confirmation. The platform provides affinity reagents for antibody-drug conjugate target assessment. The method supplies reagents for receptor internalization and functional assays.
- Companion diagnostics and precision medicine: The approach supports stratification biomarkers. The data assist in defining patient cohorts for targeted therapies. The method integrates with genomic and transcriptomic datasets.
- Imaging and targeted delivery: Aptamers conjugated to nanoparticles enable targeted imaging. The platform develops probes for cell-type selective delivery. The reagents can enhance contrast and specificity in preclinical models.
Sample Requirements
| Sample Types | Cell lines, primary cells, tissue samples (fresh or frozen), plasma, serum, or purified proteins. Fixed cells can also be used, but prior discussion is required. |
| Sample Quantity | Typically, 1–5 million viable cells for cell-based selection; 100–200 µL of serum or plasma; 50–100 µg of purified protein. Exact amounts will be confirmed during project planning. |
| Sample Quality | Cells should be viable and free from contamination. Tissues should be fresh or snap-frozen. Protein samples should be highly pure, with minimal degradation. High-quality input improves aptamer selection success. |
| Storage & Shipping | Samples should be shipped on dry ice (for frozen tissues/proteins) or in temperature-controlled containers (for cells). Creative Proteomics provides detailed packing and shipping instructions. |
Why Choose Creative Proteomics for Aptamer-Based Proteomics Profiling
- Cutting-Edge Platforms: State-of-the-art aptamer arrays and detection systems.
- Expert Scientific Team: Highly experienced specialists with molecular biology, proteomics, and bioinformatics backgrounds.
- Comprehensive Solutions: End-to-end services from sample preparation to data interpretation.
- Quality Assurance: Strict adherence to international standards for reproducibility and accuracy.
- Client-Centered Support: Dedicated project managers ensure seamless communication and tailored solutions.
FAQ
-
Q1: How does aptamer specificity compare to monoclonal antibodies?
A1: Aptamers can achieve high specificity comparable to monoclonal antibodies. Unlike antibodies, aptamers can be chemically optimized to reduce off-target interactions and can discriminate between highly similar protein isoforms or post-translationally modified variants.
-
Q2: Can aptamer-based profiling detect post-translational modifications (PTMs)?
A2: Yes. By combining aptamer enrichment with mass spectrometry, the method can identify PTMs such as phosphorylation, glycosylation, or acetylation. Aptamers can selectively capture proteins in their native state, preserving modification patterns for downstream analysis.
-
Q3: Are aptamers compatible with in vivo applications?
A3: Certain aptamers selected via in vivo SELEX can function within living organisms, targeting tissues or cell types in their native environment. Chemical modifications enhance stability and bioavailability for in vivo studies.
-
Q4: How does aptamer-based profiling integrate with other omics approaches?
A4: Aptamer-based proteomics complements transcriptomics, genomics, and metabolomics. For example, aptamer-enriched proteins can be subjected to mass spectrometry, providing proteomic data that integrates with gene expression profiles, facilitating multi-omics biomarker discovery and mechanistic studies.
Demo
Demo: Alkaline phosphatase ALPPL-2 is a novel pancreatic carcinoma-associated protein
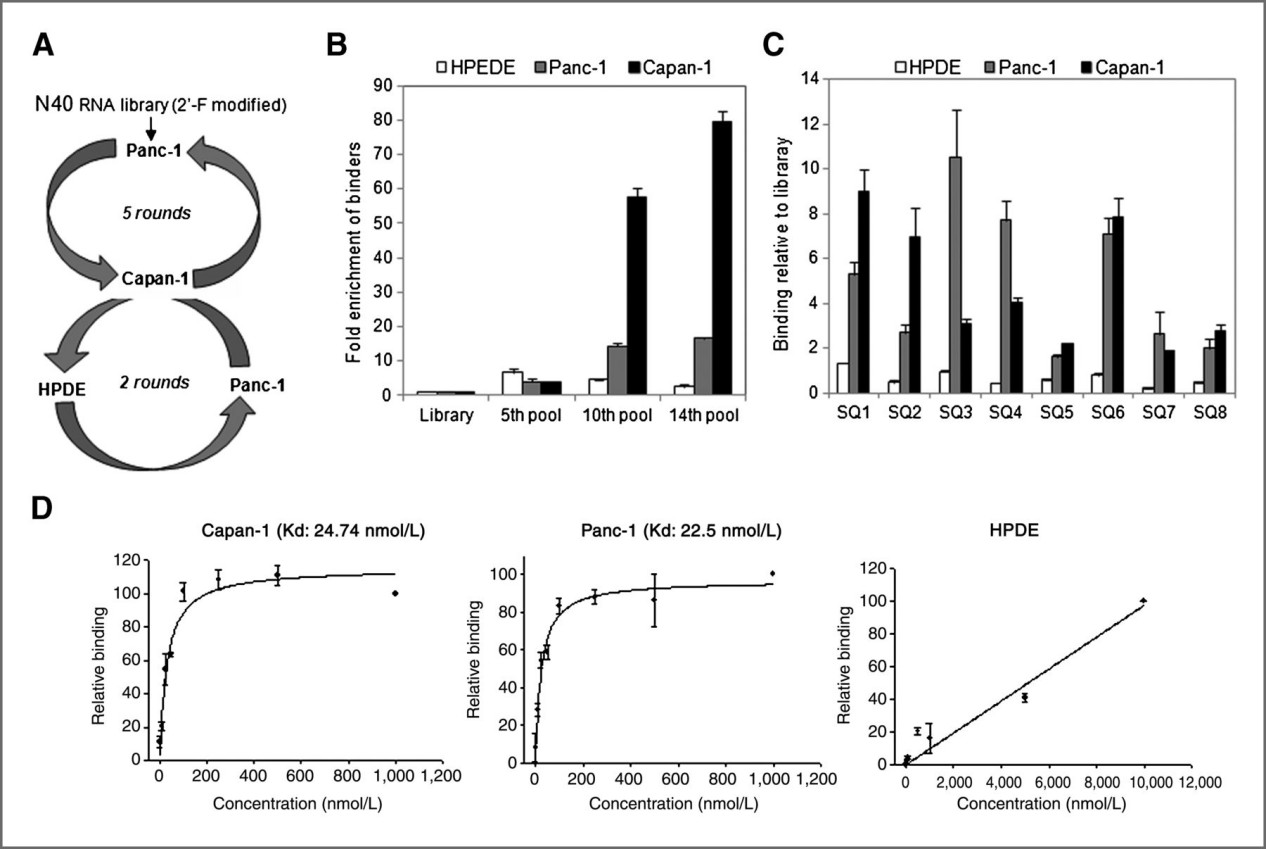
Figure 2. Cell-SELEX based identification of PDAC specific aptamer SQ-2 (Dua P, et al., 2013).
-
Case Study
Case: Aptamer-Based Proteomic Profiling Reveals Novel Candidate Biomarkers and Pathways in Cardiovascular Disease.
Abstract
Single-stranded DNA aptamers can bind proteins with high specificity and affinity. Aptamer-based multiplexed platforms enable high-throughput measurement of large numbers of low-abundance circulating proteins and thus complement traditional proteomic approaches when profiling perturbational or cohort studies. The authors aimed to assess whether an aptamer-based proteomic platform (measuring ~1,129 proteins) could detect early, dynamic protein changes after myocardial injury and identify candidate biomarkers and pathways relevant to cardiovascular disease. They also evaluated the platform's performance in epidemiologic cohorts and developed an orthogonal workflow integrating aptamer affinity capture with mass spectrometry for analytical validation.
Methods
- Sample collection and processing: Venous blood was collected into anticoagulant tubes according to a standardized protocol. Plasma was separated by centrifugation within two hours of collection. Aliquots were stored at −80 °C until analysis.
- Aptamer-based proteomic measurement: Plasma samples were analyzed using a high-plex aptamer-based proteomic assay targeting approximately 1,129 proteins.
- Aptamer affinity enrichment and mass spectrometry validation: Bound proteins were eluted, denatured, reduced, alkylated, and digested with trypsin. Peptide mixtures were analyzed by liquid chromatography–tandem mass spectrometry (LC–MS/MS). Mass spectra were searched against a human protein database.
- Analytical validation and orthogonal assays: Affinity and kinetic measurements were obtained for selected aptamers using surface-based binding assays. Concordance between aptamer-derived signals and orthogonal measurements was assessed.
Results
- 217 proteins changed significantly after planned myocardial infarction in the derivation cohort; 79 were validated in the independent cohort. More than 85% of validated proteins were directionally consistent.
- Several proteins not previously linked to acute myocardial injury were identified (for example, Dickkopf-related protein 4 and cripto), with substantial early increases in circulation. A subset of proteins that rose within 1 hour after planned injury was also elevated in spontaneous myocardial infarction cases.
- Analyses in the Framingham Heart Study found many proteins associated with the Framingham Risk Score and its components, demonstrating applicability in large cohort studies.
- The authors successfully demonstrated a workflow that couples DNA-based immunoaffinity enrichment to LC-MS/MS, providing orthogonal analytical confirmation of aptamer targets.
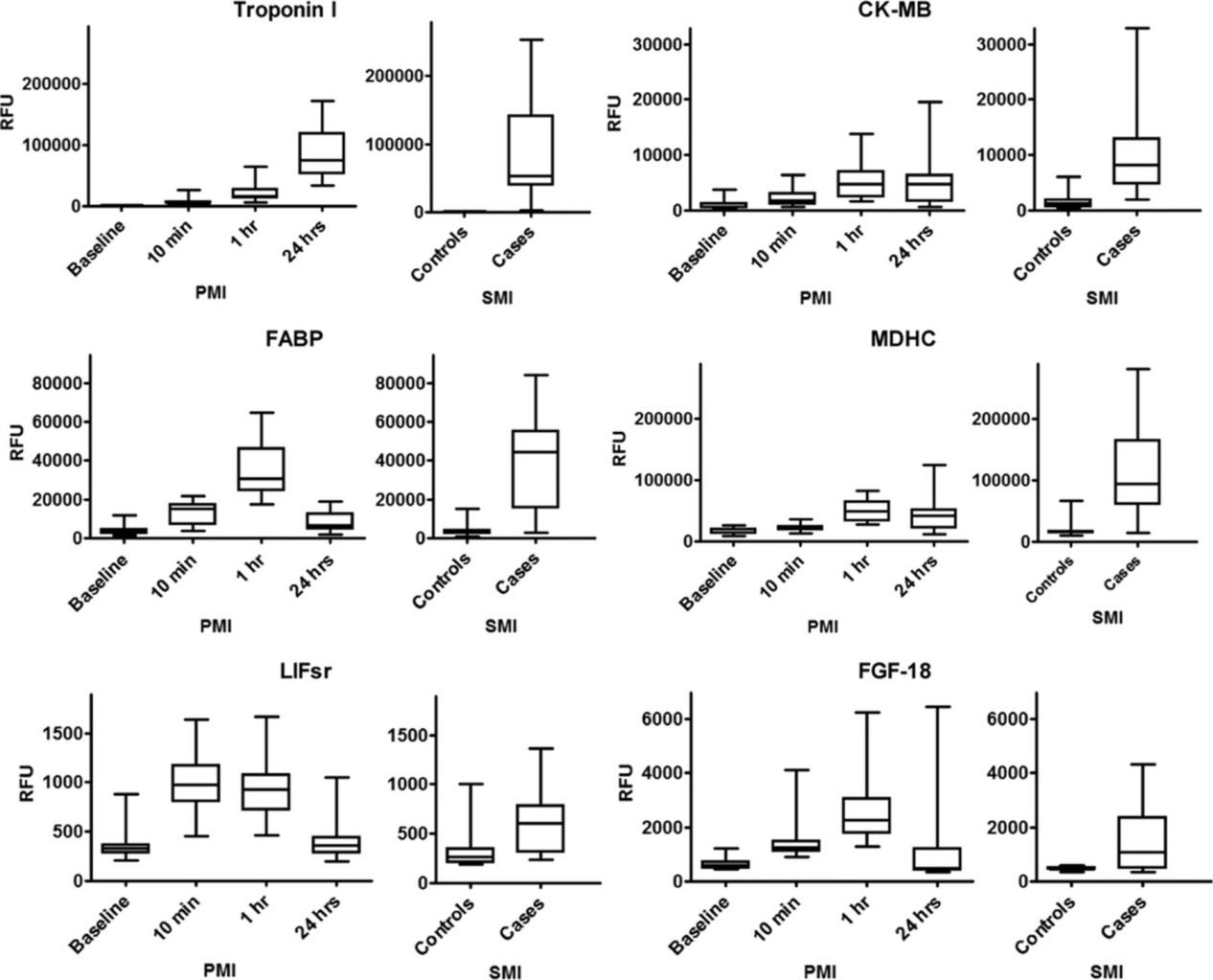
Figure 3. Proteins increased during planned and spontaneous myocardial injury.
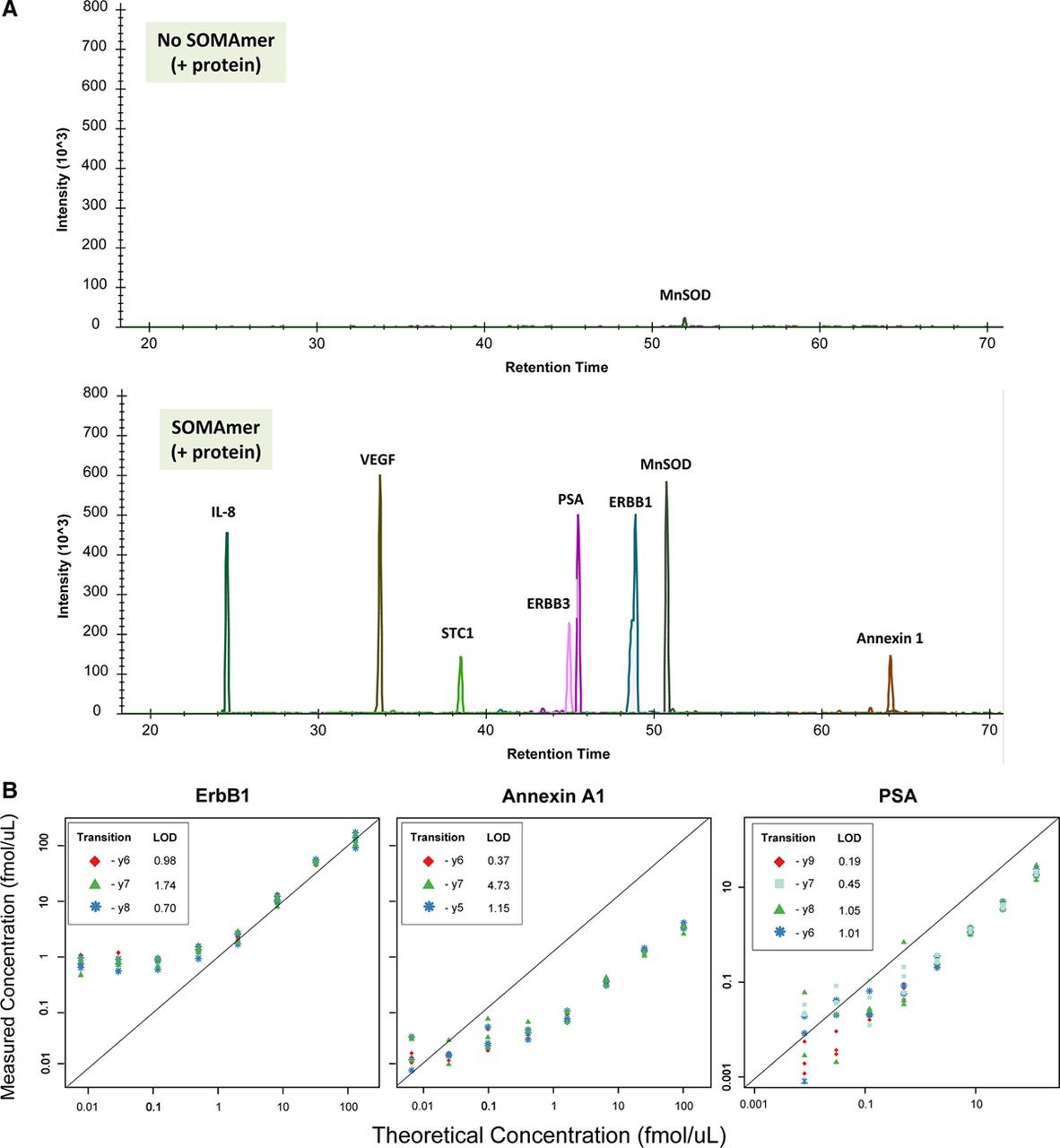
Figure 4. Mass spectrometry verification of aptamer-protein binding.
Conclusion
The study demonstrates that aptamer-based proteomic profiling is a sensitive, high-throughput approach that measures>1,000 low-abundance analytes. It can identify early protein changes after acute cardiac injury, discover novel candidate biomarkers and pathways, and scale to large cohorts. Integrating aptamer affinity enrichment with mass spectrometry provides a practical route for target validation. The platform shows promise for biomarker discovery and translational cardiovascular research.
Related Services
References
- Huang J, et al. Advances in aptamer-based biomarker discovery. Frontiers in Cell and Developmental Biology, 2021, 9: 659760.
- Ohuchi S. Cell-SELEX technology. BioResearch open access, 2012, 1(6): 265-272.
- Dua P, et al. Alkaline phosphatase ALPPL-2 is a novel pancreatic carcinoma-associated protein. Cancer research, 2013, 73(6): 1934-1945.
- Ngo D, et al. Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation, 2016, 134(4): 270-285.