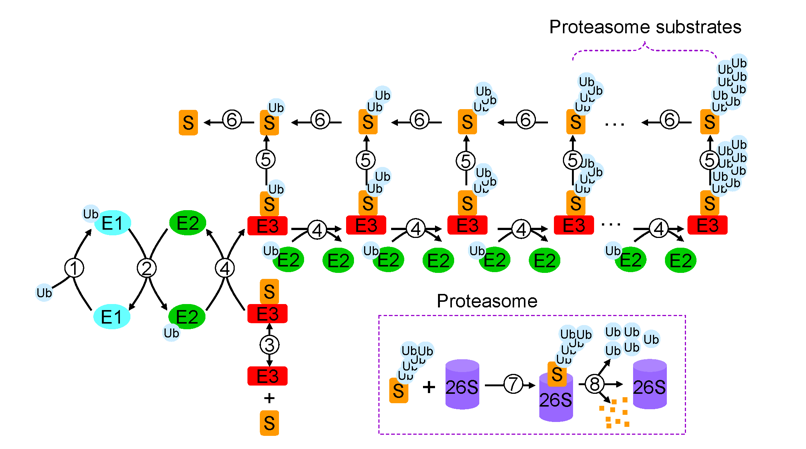
Ubiquitination is one of the important enzymatic post translational modifications occurring in eukaryotic cells. The ubiquitinated proteins contain multiple lysine residues in the structure. One or more of these lysine sites can be modified by the protein ubiquitin at the same time, resulting in macro-inhomogeneity of post-translational modification of proteins. Even at the same modification site, lysine modified homogeneous ubiquitin chains or mixed ubiquitin chains can be formed. The N-terminus of the ubiquitin molecule (the free amino group of methionine) can also undergo ubiquitination modification to form a linear ubiquitination chain, resulting in microscopic heterogeneity in complex and diverse protein ubiquitination modifications. These all reflect the diversity in structures of protein ubiquitination modification, the complexity of regulation and the importance of function in eukaryotic cells from different aspects.
The Discovery of Ubiquitin
Ubiquitin was discovered in the thymus when it was isolated from thymosin, and is considered to be an immunogenic peptide UBIP that is ubiquitous in eukaryotes. Unlike thymogenin, which only promotes the differentiation of T cells, ubiquitin can simultaneously promote the differentiation of T cells and B cells. Subsequently, Goldknopf and Busch found that the C-terminus of ubiquitin can be covalently linked to the ε-amino group of histone H2A lysine residues, revealing that ubiquitin can modify the properties of other proteins. In 1978, Hershko, Ciechanover and their colleagues discovered that the cell-free lysate of rabbit reticulocytes has the function of degrading certain substrate proteins dependent on ATP. Subsequently, Hough et al. successfully purified 26S proteasome capable of degrading polyubiquitin-protein conjugate from this rabbit reticulocyte lysate, and perfected the ubiquitin-proteasome system theory. The important role and molecular mechanism of ubiquitin-proteasome in a series of physiological processes were elucidated by Finely and his collaborators. Ubiquitin can modify the substrate protein and use it as a signal to guide the substrate to the proteasome for specific degradation, thereby maintaining the steady state of the environment in the body.
Protein Structure of Ubiquitin
Ubiquitin exists in a special form with an alpha helix surrounded by five beta sheets. This structure of SSSSSS is also present in ubiquitin-like proteins. There are several important hydrophobic surfaces on the surface of the ubiquitin molecule that are indispensable for its biological function. Among them, the hydrophobic pocket composed of Ile44, Leu8, Val70 and His68 mediates the binding of ubiquitin monomers to the proteasome and most ubiquitin binding domains (UBDs). These conserved sequences are necessary for cell division and proteasome degradation. Centered on Ile36, the hydrophobic surface formed by Leu71 and Leu73 mediates the interaction with the ubiquitin chain and the recognition of HECT-like ubiquitin ligase, deubiquitinase and ubiquitin binding domains. The surface composed of Gln2, Phe4 and Thr12 may be involved in transport and interaction with ubiquitin-specific protease domain and UBAN domain. The TEK-box composed of Thr12, Thr14, Glu34, Lys6 and Lys11 is indispensable for the degradation of mitotic-related proteins.
Ubiquitin-Proteasome and Human Health
Protein ubiquitination modification plays an important role in cell life activities such as cell division, differentiation, formation of neuronal network morphology, cell surface receptor regulation, DNA repair, transcription regulation and gene silencing, immune regulation, and inflammatory response. Disrupting any of these processes can cause a variety of serious human diseases such as Parkinson's disease, Alzheimer's disease, Huntington's disease, Prieon's disease, and amyotrophic lateral sclerosis.
Uba6 (E1) -Use1 (E2) mediates the cascade of ubiquitination of substrate proteins, which can effectively regulate the development of mouse embryos by regulating the specific degradation of these proteins. For example, the lack of Uba6 during the development of mouse embryos will cause changes in the structure of the hippocampus and amygdala neurons, a reduction in dendritic spines density and a variety of behavioral disorders.
The importance of ubiquitin as a signaling molecule has received more attention. In terms of the technology for ubiquitinated protein research, the rapid development of high-coverage quantitative proteomics technology provides an opportunity to in-depth study the ubiquitin-proteasome system. Strategies such as antibodies, epitope tags, tandem affinity purification, and site-directed mutation make it possible to enrich low-abundance ubiquitinated protein substrates. In terms of data analysis, the maturity of bioinformatics analysis has accelerated the study of protein ubiquitination in systems biology. In addition, the feasibility of using key proteins of the ubiquitin-proteasome system as drug targets has also received widespread attention.
Creative Proteomics can provide comprehensive protein ubiquitination analysis services, including protein extraction, digestion, peptide enrichment, LC-MS / MS analysis, data analysis and bioinformatics analysis. The variety of protein modification changes to meet the needs of label identification and drug target screening methods.
Reference:
- Xu L, Qu Z. Roles of protein ubiquitination and degradation kinetics in biological oscillations. PloS one, 2012, 7(4).