The definition and principle of peptide mapping
The molecular weight and amino acid composition of the proteins and peptides, as well as the specific endonucleases to act on specific peptide chain, are used in Peptide mapping. Peptides are cut into small fragments by endonucleases. A characteristic fingerprint is formed by certain means of separation and detection. Peptide mapping is helpful in structural research and peptide characterization.
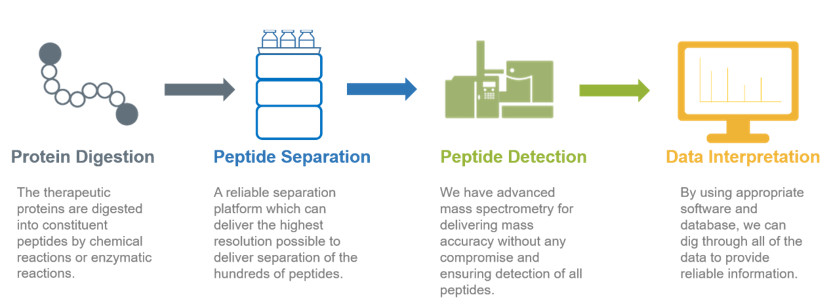
The procedure of peptide mapping
1. Obtain protein samples
2. Use 6 proteases (Trypsin, Chymotrypsin, Asp-N, Glu-C, Lys-C, and Lys-N) to digest and identify target proteins.
Using trypsin to digest the protein during the process of protein sample identification, the identified peptide coverage is about 60%. In order to obtain the entire sequence information of the target protein, six proteases (Trypsin, Chymotrypsin, Asp-N, Glu-C, Lys-C, and Lys-N) commonly used in protein identification can be selected for enzyme digestion and Identification. 100% of the protein sequence can be determined by splicing between peptides.
3. Analyze the samples using nano-flow UPLC-MS / MS.
In a peptide map, each peptide must be completely separated into a single peak. However, proteolysis products are very complicated, and appropriate chromatography methods and instruments need to be selected to separate the target products. Creative Proteomics uses ultra-performance liquid chromatography (UPLC) to analyze the peptides. Compared with high performance liquid chromatography (HPLC), UPLC has better resolution and sensitivity, and better peptide mapping capability. The small particle packing contributes to the high resolution of UPLC, producing ultra-narrow chromatography peaks. Similarly, the surface chemistry of the filling material used in the peptide separation can result in the production of excellent peak shapes.
Creative Proteomics mainly uses MALDI-TOF mass spectrometry to analyze the purified peptides. To perform MALDI TOF MS, samples are mixed with an equal amount of the matrix solution, or place the solution on the target sample. After the solvent evaporates, co-crystals of the sample and matrix are formed. Crystals are radiated using a laser as an energy source. The matrix absorbs energy from the laser to adsorb the sample. The charge transferring between the matrix and the sample ionizes the sample molecules. The sample ions enter the time-of-flight mass spectrometer for mass analysis to form a mass spectrum. Through software analysis and comparison, specific fingerprints are screened and determined. When MALDI-TOF analyzes peptide mixtures, it can tolerate appropriate amounts of buffers and salts, and almost every peptide fragment produces only single-charged ions. There is a one-to-one correspondence between the ions in the mass spectrum and the masses of the peptides and proteins, which helps the mass spectrum data analysis.
4. The complete sequence of peptide coverage/peptide spectrum is compared with the provided theoretical sequence before acquiring the final comparison result.
The application of peptide mapping
- Widely used in the quality control of genetic engineering drugs
- Amino acid structure analysis of recombinant monoclonal antibody drugs
- Identification of biological drugs.
- Study on similarity and quality of the original drug
- Identify the integrity and accuracy of the recombinant protein structure
References
1. Moore R, Samonig M. High-throughput, high-resolution peptide maps. Genetic Engineering & Biotechnology News, 2016, 36(12): 20-21.
2. Dada O O, Zhao Y, et al. High-Resolution Capillary Zone Electrophoresis with Mass Spectrometry Peptide Mapping of Therapeutic Proteins: Peptide Recovery and Post-translational Modification Analysis in Monoclonal Antibodies and Antibody–Drug Conjugates. Analytical chemistry, 2017, 89(21): 11236-11242.