Metaproteomics is a study used to qualitatively and quantitively analyze proteins in microbial communities, thus revealing microbial phenotypic information at the molecular level. The development of metaproteomics has been facilitated by liquid chromatography capable of separating highly complex peptide mixtures, high-resolution mass spectrometers capable of obtaining large amounts of precise mass spectrometric information, and computer tools capable of processing and analyzing complex data. And using liquid chromatography-tandem mass spectrometry (LC-MS/MS), tens of thousands of peptides can be identified and quantified.
From metagenomics to metaproteomics
The gut is a complex ecosystem. High-throughput metagenomic research methods, such as 16S rRNA sequencing, are widely used to study the composition of microbial communities and their impact on human health and disease. Studies have shown that these microbial communities are closely related to host resistance to pathogen infection, innate immune regulation, inflammatory response, nutrient uptake, and energy balance through metabolic processes. Imbalanced microbial communities often lead to cardiovascular and cerebrovascular diseases such as obesity or diabetes, and even atherosclerosis.
Classical microbiome studies do not help us to understand the relationship of these bacterial communities with the host genome and their impact on host health. The emergence of metaproteomics is helpful in bridging this gap.
What can metaproteomics tell us?
Compared with metagenomics and metatranscriptomics, the main advantage of metaproteomics is that it is based on "functional" information, and it is effective to study the spatiotemporal characterization of the microbial community. With the help of an accurate database of protein sequences, we can know which species (or higher taxa, such as genus, family, etc.) these proteins belong to and further understand the functional roles and interactions of different members of the community. Therefore, metaproteomics has become an important area of proteomic research to enable large-scale protein identification and analysis of microbial populations in the environment, such as the human gut.
Protein is a direct effector to maintain cell function. Detection of protein abundance is helpful to interpret cell phenotypic information at the molecular level. In addition to identifying changes in the expression of individual genes in different artificial or natural environments, metaproteomics data can also be used to analyze community structure by calculating the biomass of each member. High-resolution mass spectrometry data generated by metaproteomics can also be used to analyze the isotope content of certain proteins and species.
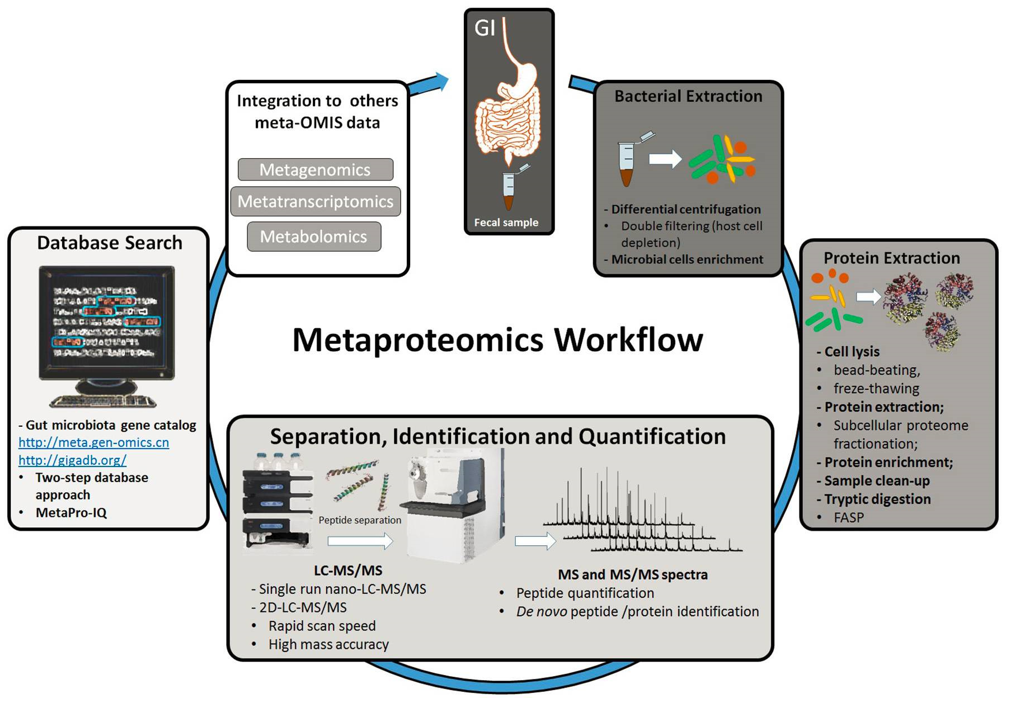
The workflow in a metaproteomic analysis of fecal sample (Petriz et al., 2017)
Metaproteomics research
The following four steps must be considered when conducting metaproteomics research: 1. Effectively extract protein from the entire microbial community; 2. Remove compounds and reagents that may interfere with restriction digestion and mass spectrometry identification; 3. Removal of host cells and enrichment of microbial cells; 4. Classification of samples before mass spectrometry detection.
Creative Proteomics can provide metaproteomics services to help you obtain microbial flora structure and function information.
Reference
Petriz, Bernardo A., and Octávio L. Franco. "Metaproteomics as a complementary approach to gut microbiota in health and disease." Frontiers in chemistry 5 (2017): 4.