Methylation can occur on Arg, lys, Asp, Asn, Gln, His, Glu and cys, or on the N terminus or C terminus of the protein. According to the position of methyl binding, there are three types of methylation, namely N-methylation, O-methylation, and S-methylation.
Histone methylation refers to the addition of one, two or three methyl groups to certain amino acids in histones, called mono-, di- and trimethylation, respectively. Although methylation can occur at many sites in histones, it occurs primarily at the trailing lysine (lysine, K) and arginine (arginine, R) residues. Histone methylation modifications are regulated by histone methyltransferases (HMTs) and histone demethylases (HDMs). Methylation of histone lysine residues can activate or repress gene transcription, depending on the specific circumstances (e.g., site of methylation, status, etc.).
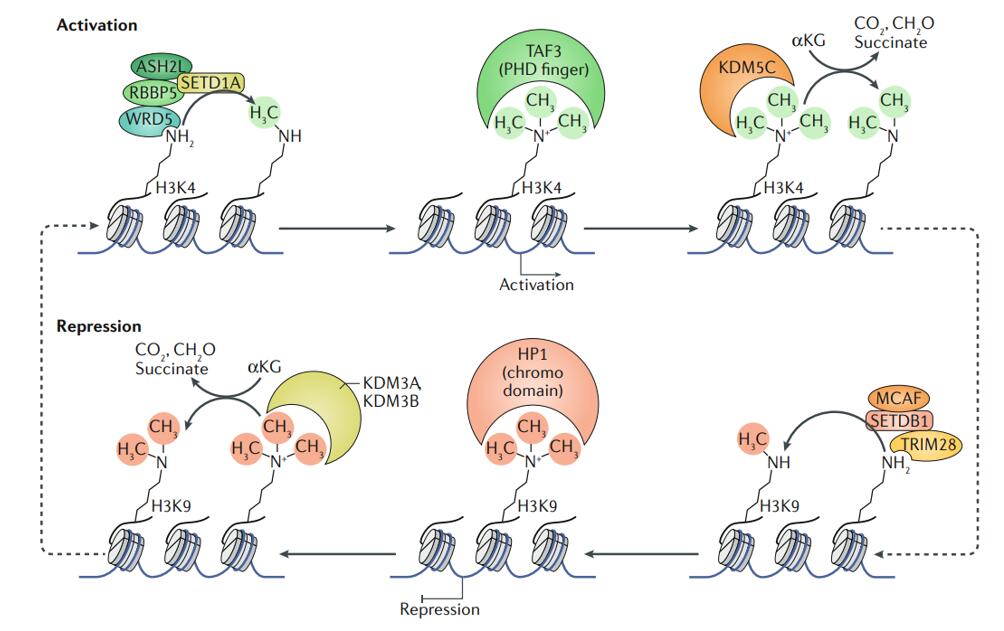
Examples of regulation of gene expression by histone methyltransferases and demethylases
(Jambhekar et al., 2019)
In the nucleus, histone methylation is closely related to a variety of life activities, and has different biological effects due to different methylation sites, mainly in heterochromatin formation, gene imprinting, X chromatin inactivation and transcriptional regulation. With the development of epigenetics, histone methylation detection technology is also advancing, and there are various histone methylation detection methods as follows.
Chromatin Immunoprecipitation (CHIP)
Nuclei are extracted and digested at 20°C using standard micrococcal nuclease. Chromatin fragments of exact length are separated by sucrose gradient centrifugation. Non-denaturing chromatin immunoprecipitation is performed by specific antibodies. Analysis of chromatin immunoprecipitation is used to study proteins with high affinity for DNA. High resolution at the mononucleosome level can be achieved, and the efficiency of immunoprecipitation can be grasped.
The proteins in cells or tissues separated by electrophoresis are transferred from the gel to a solid phase support NC membrane or PVDF membrane, and then a specific antigen is detected with a specific antibody. This method can only detect methylated histones at the overall level and cannot meet the deeper needs.
Biochip
Commonly used biochips include gene chip and protein chip, which can provide histone methylation expression profiles covering the whole genome sequence sites and help researchers to select histone methylation sites with research significance.
Mass Spectrometry Analysis
Methylation modifications, similar to other post-translational modifications, are in low abundance in organisms. If identified directly by enzymatic digestion, the signal may be overwhelmed and difficult to detect. Creative Proteomics has developed a protein methylation modification analysis platform that allows for the enrichment of specific methylation modifications that can then be detected by mass spectrometry. The HPLC-MS/MS based platform can identify, quantify and characterize protein methylation. Methylation site analysis and methylation proteomics services can be provided.
Reference
[1] Jambhekar, A., Dhall, A., & Shi, Y. (2019). Roles and regulation of histone methylation in animal development. Nature reviews Molecular cell biology, 20(10), 625-641.