Acetylation refers to a reaction that introduces an acetyl functional group into a chemical compound, in which the hydrogen atom of a hydroxyl group is replaced by an acetyl group (CH3 CO) to yield a specific ester, the acetate. Protein acetylation commonly has two different forms. In humans, almost (80%-90%) proteins become co-translationally acetylated at their Nα-termini of the nascent polypeptide chains. Another type is typically acetylated on lysine residues.
N-terminal acetylation
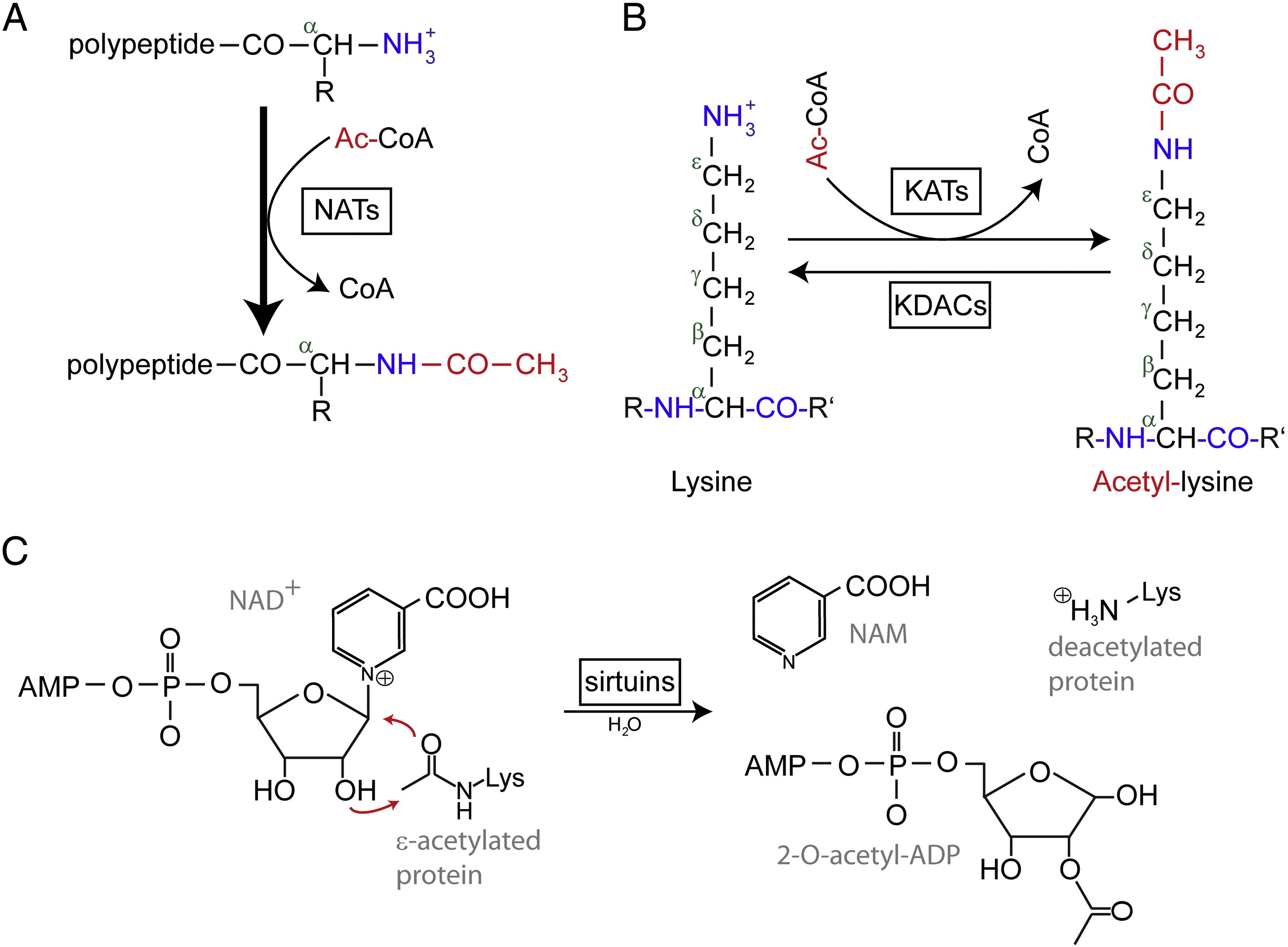
N-terminal (Nt) acetylation are catalyzed by Nt-acetyltransferases (NATs) and is found to be irreversible so far. NATs, mono- or multisubunit enzymes consisting of a catalytic subunit and up to two auxiliary subunits, can transfer an acetyl group from acetyl-coenzyme A (Ac-CoA) to the α-amino group of the first amino acid residue of the protein. In NATs, the major auxiliary subunit modulates the activity and substrate specificity of the catalytic subunit. Different NATs are responsible for the Nt acetylation. In humans, there are six NATs have been found so far, including NatA, NatB, NatC, NatD, NatE, and NatF. In addition to the difference in subunit composition, the various NATs vary in their substrate specificities.
Nt acetylation plays different roles in molecular effects. Firstly, Nt-acetylation determines the subcellular localization for certain proteins. For example, Arl3 and Grh1, two Golgi-associated proteins, cannot associate with the Golgi apparatus when missing the Nt-acetyl group. Secondly, it is reported that Nt-acetylation restrains proteins in the cytosol and inhibits a post-translational translocation migration to the endoplasmic reticulum (ER) and the secretory pathway. In addition, Nt acetylation can alter the properties of the N-terminus to make protein-protein interactions become modulated. It was shown for several proteins that the affinity to their binding partners increased after being Nt-acetylated. For example, The E2 ubiquitin-conjugating enzyme Ubc12 undergoes Nt-acetylation by NatC enabling an increased affinity towards its interaction partner, the E3 ubiquitin ligase Dcn1. Moreover, Nt-acetylation controls protein quality and lifetime, and regulates the protein stoichiometry by the N-end rule pathway.
N-terminal acetylation has many functions in physiology. NATs are essential for normal development, bone and blood vessel development. N-terminal acetylation can regulate blood pressure, proteasome localization, hormone, as well as organelle structure and function. In human disease, it related to neurodegenerative diseases (such as Alzheimer's disease, Parkinson's disease, and Lewy body dementia) and cancer (like lung cancer, breast cancer, colorectal cancer).
Lysine acetylation
Acetylated lysine residues were first discovered in histones regulating gene transcription. But lysine acetylation is not limited to histones. Unlike Nt acetylation, lysine acetylation is reversible. The acetylation is catalyzed by lysine acetyltransferases (KATs) and the deacetylation of lysine residues is catalyzed by Lysine deacetylases (KDACs).
It is reported that 17-22 genes KATs have been identified in the human genome (The exact number of KATs is controversial), which can be classified into three different families, including GCN5 (general control non-derepressible 5)-related acetyltransferase (GNAT) family, the MYST family, and p300/CBP (CREB-binding protein) family. The known substrates of KAT complexes not only include histone proteins, but some different transcription factors, transcriptional co-regulators, and some proteins of specific cellular signaling pathways like p53, β-catenin, NF-κB, MyoD or Rb. And even some RNA molecules can be acetylated. There are four different types of KDACs, including Class I, II, III, and IV. Class I, II and IV are Zn2 +-dependent amidohydrolases, whereas class III (also called sirtuins) uses NAD+ as co-substrate for its catalytic activity are Zn2 +-dependent amidohydrolases.
The histones are the first discovered acetylated proteins. The histones molecules are modified by different PTMs, including phosphorylation, methylation, and acetylation. histone acetylation, determine the histone assembling as well as the folding and compactness of the DNA-histone interaction and therefore presenting a switch between permissive and repressive chromatin structure. In addition to histones, KATs can catalyze cytoskeletal proteins. There are some novel identified acetylation sites in other proteins, including HMG proteins, c-Myc, estrogen and androgen receptors, E2F/Rb and so on.
The physiological roles of lysine acetylation have been reported. The histone proteins are associated with a tight regulation of essentially all types of DNA-templated processes as transcription, replication, recombination, repair, as well as translation and formation of specialized chromatin structures. Therefore, protein lysine acetylation affects a range of cellular signaling pathways as well as metabolism, stress responses, apoptosis, and membrane trafficking.
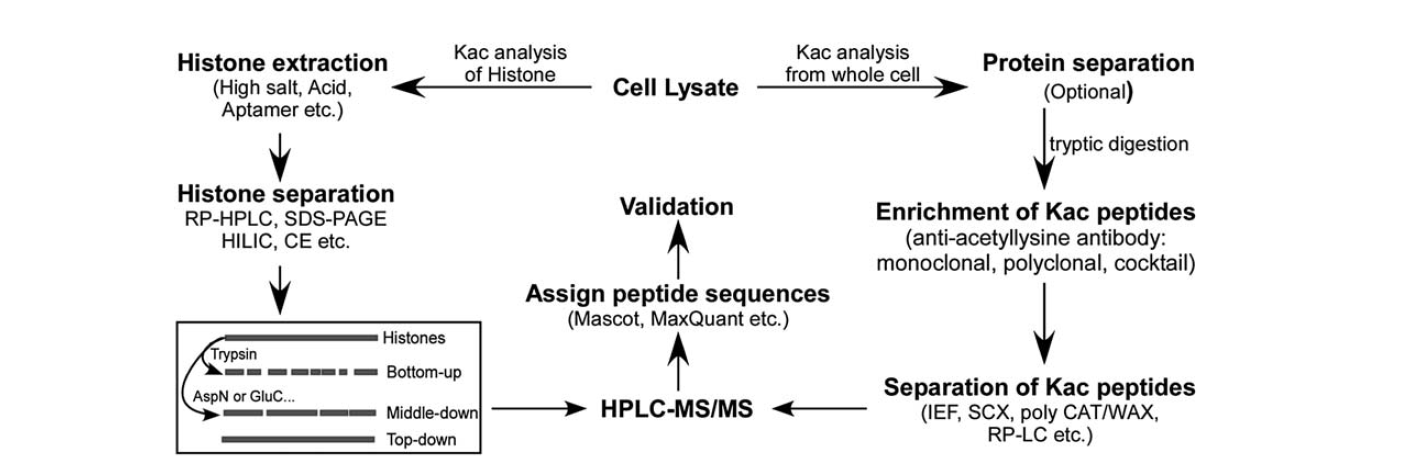
There are relatively fewer tools available for identifying lysine acetylation. With the development of mass spectrometry, it has been an important tool for the identification and quantification of lysine acetylation. To analysis histone lysine acetylation, there are several steps, including cell lysate, histone extraction, histone separation, mass spectrometry, and data analysis. The steps for analysis for protein lysine acetylation from the whole cell include cell lysate, protein separation, enrichment of lysine acetylation peptides, separation of lysine acetylation peptides, mass spectrometry, and data analysis.
References:
- Drazic, Adrian, et al. "The world of protein acetylation." Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics1864.10 (2016): 1372-1401.
- Zhang, Kai, Shanshan Tian, and Enguo Fan. "Protein lysine acetylation analysis: current MS-based proteomic technologies." Analyst138.6 (2013): 1628-1636.