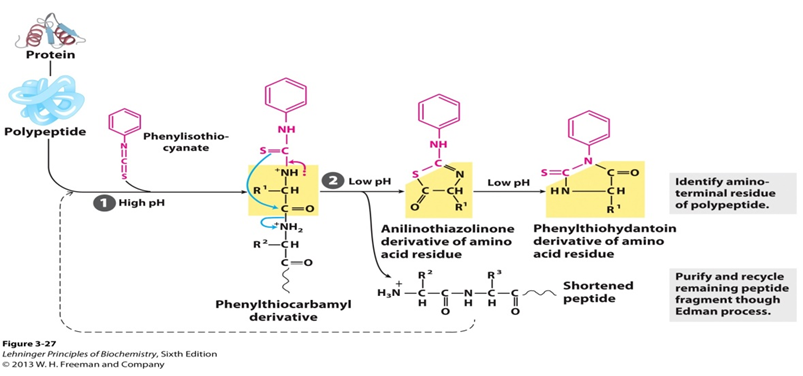
The sequence of amino acids in a protein or peptide can be identified by Edman degradation, which was developed by Pehr Edman. This method can label and cleave the peptide from N-terminal without disrupting the peptide bonds between other amino acid residues. The Edman degradation reaction was automated in 1967 by Edman and Beggs. Nowadays, the automated Edman degradation (the protein sequenator) is used widely, and it can sequence peptides up to 50 amino acids.
Cyclic degradation of peptides based on the reaction of phenylisothiocyanate with the free amino group of the N-terminal residue such that amino acids are removed one at a time and identified as their phenylthiohydantoin derivatives. Speaking to the specific process, an uncharged peptide is reacted with phenylisothiocyanate (PITC) at the amino terminus under mildly alkaline conditions to give a phenylthiocarbamoyl derivative (PTC-peptide). Then, under acidic conditions, the thiocarbonyl sulfur of the derivative attacks the carbonyl carbon of the N-terminal amino acid. The first amino acid is cleaved as anilinothiazolinone derivative (ATZ-amino acid) and the remainder of the peptide can be isolated and subjected to the next degradation cycle. Once formed, this thiazolone derivative is more stable than phenylthiocarbamyl derivative. The ATZ amino acid is then removed by extraction with ethyl acerate and converted to a phenylthiohydantoin derivative (PTH-amino acid). And the chromatography can be used to identify the PTH residue generated by each cycle.
As to the automated Edman degradation, proteins can be analyzed by applying them in-solution onto a TFA filter and then loaded onto the Edman sequencing instrument. Proteins in mixtures are first separated by 1D or 2D gels and then blotted onto a PVDF membrane. The proteins are detected by Coomassie blue, Amido black or Poncau S staining and the proteins of interest cut out and the PVDF membrane piece loaded onto the Edman sequencer.
With mass spectrometry was developed, the use of Edman degradation sequencing began to decrease. However, it stills remains the methods for several types of protein structural analysis applications. It can be used to verify the N-terminal boundary of recombinant proteins or determining the N-terminus of protease-resistant domains, particularly when the protein or domain is >40 to 80 kDa or cannot be readily purified. It also can be used to identify the new N-terminal and proteolytic cleavage site in the protein fragments. In addition, as to some novel proteins and peptides where sequence databases are not available for MS/MS database searching, Edman degradation can be used for analysis.
The N-terminal amino acid of the protein can be cleaved off. Thus, in the process, the first cycle thus identifies the exact N-terminal amino acid. In addition, because the released amino acids are identified and quantified by chromatography, the amino acids with identical molecular weight can be identified. For example, isoleucine and leucine have a mass of 113 Da, but they have different retention time. Moreover, Edman sequencing can be performed on PVDF blots from 1D and 2D gels, which enables N-terminal sequencing of proteins in the mixture. However, Edman degradation sequencing will not be available when the peptide whose N-terminus has been chemically modified, such as acetylation. And as the PITC cannot reactive with non-α-amino acid, Sequencing will stop if a non-α-amino acid is encountered like isoaspartic acid. Moreover, larger proteins cannot be sequenced by the Edman sequencing.
At Creative Proteomics, our professional protein sequencing platform provides N-terminal sequence analysis by Edman degradation or mass spectrometry (MS), with complementary advantages to each other. Our team of experts with extensive experience can help you understand what you are trying to investigate and meet your requirements. In addition, we can provide other protein identification services, including:
- Peptide mass fingerprinting (PMF) service
- Sequence analysis of peptides or proteins
- De novo protein sequence analysis service
- Shotgun protein identification service
- Membrane proteomics service
References:
1. Smith J B. Peptide sequencing by Edman degradation. eLS, 2001.
2. Reim D F, Speicher D W. N‐Terminal Sequence Analysis of Proteins and Peptides. Current protocols in protein science, 2001: 11.10. 1-11.10. 38.