Mass spectrometry is an analytical technique in which samples are ionized into charged molecules and ratio of their mass-to-charge (m/z) can be measured. In MALDI-TOF mass spectrometry, the ion source is matrix-assisted laser desorption/ionization (MALDI), and the mass analyzer is time-of-flight (TOF) analyzer.
MALDI
MALDI is a soft ionization that involves a laser striking a matrix of small molecules to make the analyte molecules into the gas phase without fragmenting or decomposing them. Some biomolecules are too large and can decompose when heated, and traditional techniques will fragment or destroy macromolecules. MALDI is appropriate to analyze biomolecules like peptides, lipids, saccharides, or other organic macromolecules.
MALDI In Omics
MALDI is hailed for its versatile applications in a multitude of omics fields, including proteomics, metabolomics, lipidomics and glycomics.
A technique that can analyze complex protein mixtures, MALDI-TOF mass spectrometry is particularly useful in determining protein molecular weight, identifying protein sequences, recognizing protein structures, and quantifying protein content.
MALDI-TOF mass spectrometry can also be employed in the realm of Glycomics, the study of glycans, particularly in glycosylation and glycan structure analysis. The technology's impressive capabilities extend to Lipidomics as well, where it can be used to identify and quantify lipid molecules, encompassing lipids, lipid metabolites, and other lipid-related molecules.
With its many diverse applications, it is clear that MALDI technology is a critical tool for the analysis and identification and quantification of metabolites including lipid-related molecules.
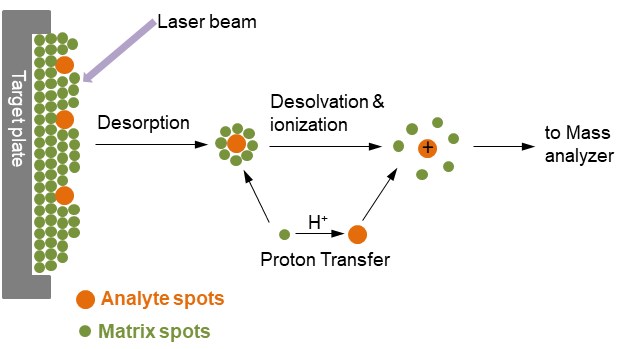
Figure 1. Ionization of analytes by MALDI
- The principle of MALDI
In Figure 1, the analyte is embedded in a very large excess of a matrix compound deposited on a solid surface called a target, usually made of a conducting metal and having spots for several different samples to be applied. After a very brief laser pulse, the irradiated spot is rapidly heated and becomes vibrationally excited. The matrix molecules energetically ablated from the surface of the sample, absorb the laser energy and carry the analyte molecules into the gas phase as well. During the ablation process, the analyte molecules are usually ionized by being protonated or deprotonated with the nearby matrix molecules. The most common MALDI ionization format is for analyte molecules to carry a single positive charge.
- Types of laser commonly used in MALDI
Lasers of both ultraviolet (UV) and infrared (IR) wavelengths are in use, but UV lasers are by far the most important light sources in analytical MALDI. Among these, nitrogen lasers and frequency-tripled or quadrupled Nd: Yag lasers often serve for the majority of applications. IR-MALDI is dominated by Er:Yag lasers while TEA-CO2 lasers are rarely used.
- Commonly used MALDI matrix substance
It is believed that the first function of the matrix essentially is to dilute and isolate analyte molecules from each other. This occurs during solvent evaporation and concomitant formation of a solid solution. Then, upon laser irradiation, it functions as a mediator for energy absorption. The choice of the right matrix is key to the success in MALDI. In general, highly polar analytes work better with highly polar matrices, and nonpolar analytes are preferably combined with nonpolar matrices. As shown in Table 1, different matrixes have been sought and widely used. Currently, the most commonly used matrixes are α-cyano-4-hydroxycinnamic acid, 2,5-dihydroxybenzoic acid, 3,5-dimethoxy-4-hydroxycinnamic acid, and 2,6-dihydroxyacetophenone.
Table 1. UV-MALDI matrices (Gross J. H., 2006)
| Compound | Acronym | Application to |
|---|---|---|
| Nicotinic acid | NA | Peptides, proteins |
| Picolinic acid | PA | Oligonucleotides, DNA |
| 3-Hydroxypicolinic acid | HPA, 3-HPA | Oligonucleotides, DNA |
| 3-Aminopicolinic acid | 3-APA | Oligonucleotides, DNA |
| 6-Aza-2-thiothymine | ATT | Oligonucleotides, DNA |
| 2,5-Dihydroxybenzoic acid | DHB | Proteins, oligosaccharides |
| DHB-based mixtures | DHB/XY and super-DHB | Proteins, oligosaccharides |
| 3-Aminoquinoline | 3-AQ | Oligosaccharides |
| α-Cyano-4-hydroxycinnamic acid | α-CHC, α-CHCA, 4-HCCA, CHCA | Peptides, smaller proteins, triacylglycerols, numerous other compounds |
| 4-Chloro-α-cyano-cinnamic acid | ClCCA | Peptides |
| 3,5-Dimethoxy-4-hydroxycinnamic acid | SA | Proteins |
| 2-(4-Hydroxyphenylazo) benzoic acid | HABA | Peptides, proteins, glycoproteins, polystyrene |
| 2-Mercaptobenzothiazole | MBT | Peptides, proteins, synthetic polymers |
| 5-Chloro-2-mercaptobenzothiazole | CMBT | Glycopeptides, phosphopeptides, and proteins |
| 2,6-Dihydroxyacetophenone | DHAP | Glycopeptides, phosphopeptides, proteins |
| 2,4,6-Trihydroxyacetophenone | THAP | Solid-supported oligonucleotides |
| Dithranol (1,8,9-anthracenetriol) | None | Synthetic polymers |
| 9-Nitroanthracene | 9-NA | Fullerenes and derivatives |
| Benzo[a]pyrene | None | Fullerenes and derivatives |
| 2-[(2E)-3-(4-tert-Butylphenyl)-2-methylprop-2-enylidene]malonitrile | DCTB | Oligomers, polymers, dendrimers, small molecules |
Time of Flight (TOF) analyzer
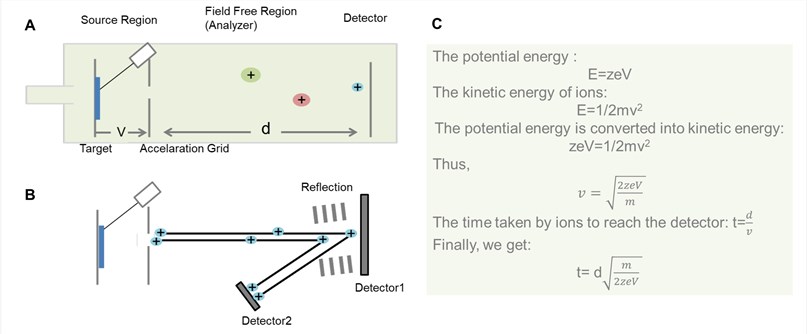
Figure 2. General schematic for TOF analyzer. (A) Liner TOF analyzer; (B) Reflector TOF analyzer; (C) The derivation process of the time that ions pass through field free region in the liner TOF analyzer.
- The principle of TOF
As shown in Figure 2, the basic principle of TOF is that ions of different m/z are dispersed in time during their flight along a field-free drift path of known length. Provided that all the ions start their journey at the same time or at least within a sufficiently short time interval, the lighter ones will arrive earlier at the detector than the heavier ones.
- Liner TOF analyzer and reflectron TOF analyzer
Theoretically, all the ions are given the same initial kinetic energy, so that after drifting along the field free region, the ions of the same m/z at the detector at the time. However, in practice, the pulse is not felt by all ions to the same intensity and so not all the ions of the same m/z values reach their ideal velocities. To correct this problem, a reflection is often applied to the end of the drift zone. The reflectron consists of a series of ring electrodes with high voltage, which can repulse the ions back along the flight tube usually at a slightly displaced angle.
Ions of different kinetic energy penetrate the reflectron to different depths before they get expelled from the reflectron into the opposite direction. Faster ions carrying more kinetic energy will travel a longer path than slower ones, and thus spend more time within the reflectron than slower ions carrying less energetic. In that way, the detector receives ions of the same mass at (about) the same time. Thereby, this design for TOF mass analyzer has increased their resolution significantly. However, reflectron TOF analyzer is not suitable for analytes that are not stable enough to survive the electric field.
The process of MALDI-TOF mass spectrometry
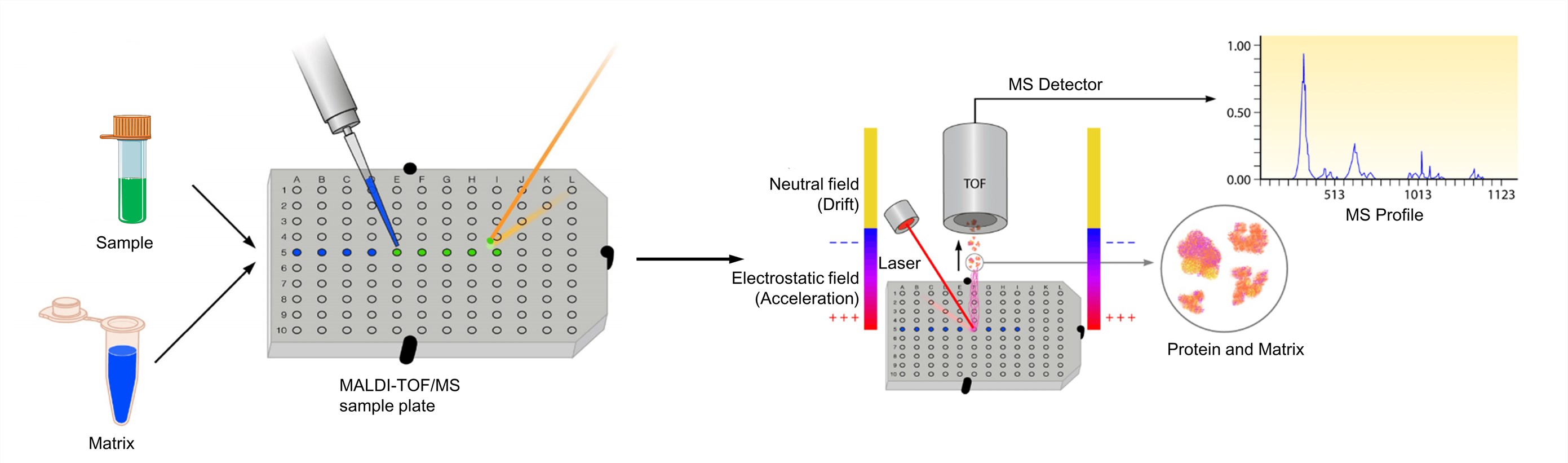
Figure 3. The process of MALDI-TOF mass spectrometry (Clark A. E., et al.; 2013)
The analyte should be soluble to at least about 0.1 mg/ml in some solvent. And the matrix is dissolved to yield either a saturated solution or a concentration of about 10 mg/ml. The solution of the analyte is then admixed to that of the matrix. For optimized MALDI spectra, the molar matrix-to-analyte ratio is normally adjusted as to fall into the range from 1000: 1 to 100,000: 1. And then the mixture is then spotted onto a metal target plate for analysis. After drying, the mixture of the sample and matrix co-crystallizes and forms a solid deposit of sample embedded into the matrix. The plate is subsequently loaded into the MALDI-TOF instrument and analyzed by software associated with the respective system. The MALDI leads to the sublimation and ionization of both the sample and matrix. These generated ions are separated depending on m/z through a TOF analyzer, and a spectral representation of these ions is generated and analyzed by the MS software, generating an MS profile.
Application of MALDI-TOF mass spectrometry
- Intact Mass determination
The intact mass determination is basic and important for protein characterization, due to the correct molecular weight of a protein can indicate the intact structure. MALDI, a soft ionization technique, is suitable for proteins which tend to be fragile and fragment when ionized by other ionization methods. The performance of MALDI-TOF MS is less affected by buffer components, detergents, and contaminants. In addition, it permits intact protein mass determination with sufficient accuracy (≤ 500 ppm) for sequence validation. After protein digestion, MALDI-TOF MS can be also used to analyze the obtained peptides for further primary sequence confirmation by peptide mass fingerprinting.
- Peptide mass fingerprinting (PMF)
MALDI-TOF mass spectrometry has simple operation, good mass accuracy, as well as high resolution and sensitivity. Therefore, it has widespread uses in proteomics to identify proteins from simple mixtures by a method called peptide mass fingerprinting, which are often used with two-dimensional gel electrophoresis (2-DE). In this approach, peptides are generated by digesting proteins of interest with a sequence-specific enzyme like trypsin. And then peptides are analyzed by MALDI-TOF mass spectrometry to get the peptide masses. The experimental masses are compared against a database containing theoretical peptide masses from a given organism with the same sequence-specific protease.
- Post source decay (PSD) MALDI-TOF analysis
MALDI-TOF mass spectrometers equipped with reflectrons can analyze fragment ions produced from precursor ions that spontaneously decompose in the flight. Such ions are generally referred to as metastable ions, and the process of decomposition in the field free region between the ion source and the reflectron is commonly referred to as PSD. PSD fragment ions are formed within the field free region before entering the reflectron. PSD fragment ions can be separated, collected, and recorded on the detector by continuously changing the reflector voltage to form a PSD mass spectrum that provides very rich and effective structural information for the primary structure of peptides and proteins. In the proteomics study, some 2DE-separated protein samples cannot be identified by PMF or the results of identification are not clear. The PSD sequencing function can be applied to the identification of these proteins. Using PSD spectroscopy, combined with a database search, proteins can be identified quickly and with high specificity.
- Oligonucleotides analysis
With the development of molecular biology techniques and antisense nucleic acid drug technologies, more and more oligonucleotide fragments have been synthesized to be used as primers, probes and antisense drugs. It is entirely necessary to quickly detect these fragments to determine whether the synthesis is complete and whether the synthesized sequence is correct. Mass spectrometry, including MALDI-TOF-MS, is by far the best means of doing this. Oligonucleotide analysis using MALDI-TOF-MS was simple, rapid, accurate, and sensitive, which can be used to determine the complete oligonucleotide sequence.
- MALDI imaging
The MALDI-TOF can be used in profiling and imaging proteins directly from thin tissue sections, known as MALDI imaging mass spectrometry (MALDI-IMS). It provides specific information about the local molecular composition, relative abundance and spatial distribution of peptides and proteins in the analyzed section. MALDI-IMS can analyze multiple unknown compounds in biological tissue sections simultaneously through a single measurement that can obtain molecule imaging of the tissue while maintaining the integrity of cells and molecules in tissues.
MALDI-TOF mass spectrometry can analyze a wide variety of biomolecules, such as peptides, proteins, carbohydrate, oligonucleotide, and so on. Due to the fact that formed ions have low internal energy, a great advantage of MALDI-TOF is that the process of soft-ionization enables observation of ionized molecules with little to fragmentation of analytes, allowing the molecular ions of analytes to be identified, even within mixtures. And it is easy to use and maintain with fast data acquisition. Choosing the appropriate matrix substance is important for successful MALDI-TOF mass spectrometry.
At Creative Proteomics, we can provide various services based on our advanced MALDI-TOF mass spectrometry platforms, including:
- Molecular Mass Determination
- Peptide Mass Fingerprinting (PMF)
- Imaging Mass Spectrometry (IMS)
References
- Gross J H. Mass spectrometry: a textbook. Springer Science & Business Media, 2006.
- Boesl U. Time - of - flight mass spectrometry: Introduction to the basics. Mass spectrometry reviews, 2017, 36(1): 86-109.
- Guerrera I C, Kleiner O. Application of mass spectrometry in proteomics. Bioscience Reports, 2005, 25(1-2): 71-93.
- Fuchs B, Schiller J. Application of MALD - TOF mass spectrometry in lipidomics. European Journal of Lipid Science and Technology, 2009, 111(1): 83-98.
- Duncan M W, Roder H, Hunsucker S W. Quantitative matrix-assisted laser desorption/ionization mass spectrometry. Briefings in functional genomics and proteomics, 2008, 7(5): 355-370.
- Kenny D J, Brown J M, Palmer M E, et al. A parallel approach to post source decay MALDI-TOF analysis. Journal of the American Society for Mass Spectrometry, 2006, 17(1): 60-66.



