- Service Details
- FAQ
- Case Study
An increasing number of studies have shown that lipids are not only skeletal components of biological membranes and energy storage substances, but also involved in many important functions of cells. Many major human diseases have been shown to be associated with disorders of lipid metabolism, such as Alzheimer's disease, diabetes, obesity, cancer, neurodegenerative diseases, and certain infectious diseases. Lipids mainly include the following eight categories: fatty acids (e.g. linoleic acid, arachidonic acid), glycerolipids (e.g. TG, DG), glycerophospholipids (e.g. PC, PE, PG, PA, etc.), sphingolipids (e.g. Cer, SM, etc.), sterolipids (e.g. sterolipids), glycolipids (e.g. MGDG, SQDG, etc.), pregnenolipids (e.g. CoA, etc.), polyethylene.
Lipidomic analysis is the systematic analysis and identification of lipids in organisms, tissues or cells and the molecules with which they interact. In addition, the structure and function of lipids can also be understood, so as to reveal the relationship between lipid metabolism and the physiological and pathological processes of cells, organs and even the body. The contents of lipidomics research mainly include analysis and identification of lipids and their metabolites, lipid function and metabolic regulation, lipid metabolism pathways and networks. Lipidomics can provide a holistic and large-scale understanding of the role played by lipids in life processes, and may identify new markers that can provide new ideas for future disease cure or drug resistance studies.
Lipidomics Service at Creative Proteomics
At Creative Proteomics, we can provide a wide range of lipidomics profiling.
- Fatty Acids Analysis Service
- Fatty Acids Derivatives Analysis
- Fatty Acids Metabolism Analysis Service
- Sphingolipids
- Sterol Lipids Analysis Service
- Glycerolipids Analysis Service
- Glycerophospholipids Analysis Service
- Other Analysis Services
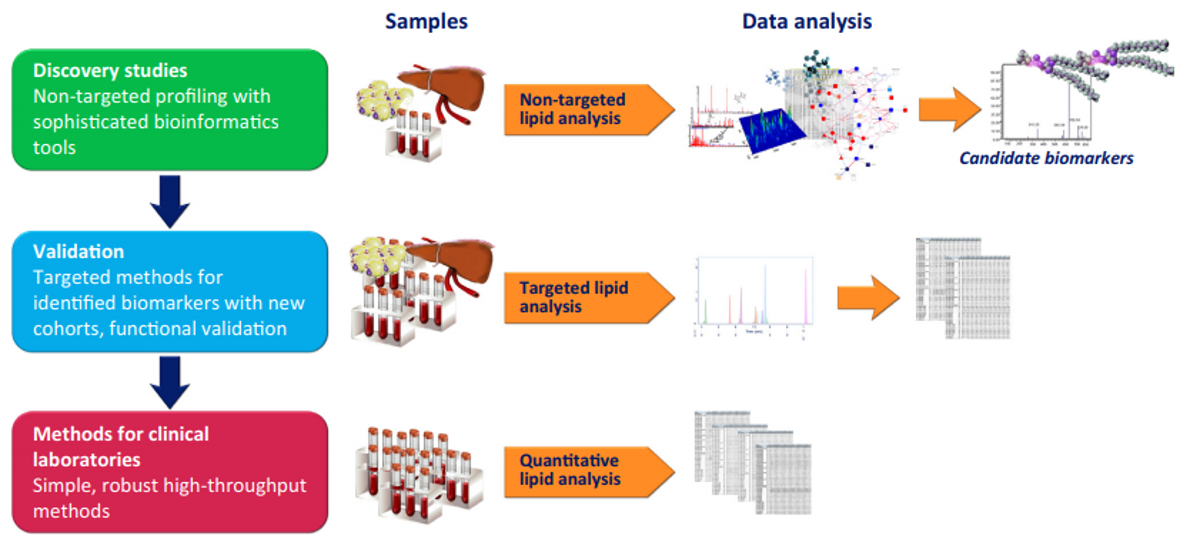 Figure 1. Analytical Workflows in Lipidomics (Hyötyläinen et al.,2015)
Figure 1. Analytical Workflows in Lipidomics (Hyötyläinen et al.,2015)
Technology Platform
The techniques of lipidomics research mainly include the extraction, separation, analysis and identification of lipids and the corresponding bioinformatics techniques. Creative Proteomics mainly provides lipidomic analysis services based on biological mass spectrometry technology. The lipid analysis strategy based on mass spectrometry mainly includes liquid chromatography-mass spectrometry and shotgun lipidomics.
- The liquid chromatography-mass spectrometry technology strategy is to use different lipid extraction methods to extract different types of lipids (lipid classes), or according to the difference in polarity of different types of lipids. Normal phase chromatography is used to classify the lipids of biological samples into different components at the species level. Then reverse phase chromatography is used to further separate the lipid molecules (molecular species) in the components, and then qualitative and quantitative analysis is performed using mass spectrometry.
- The shotgun lipidomics technique strategy usually adopts direct injection, and the lipid extract is directly analyzed and identified without chromatographic separation.
- AB Triple TOF 6600(Agilent 1290 UHPLC-AB 6600 MS)
- ScientificTM Orbitrap FusionTM LumosTM TribridTMTriple UHPLC-Mass Spectrometer System
- Orbitrap Mass Spectrometer
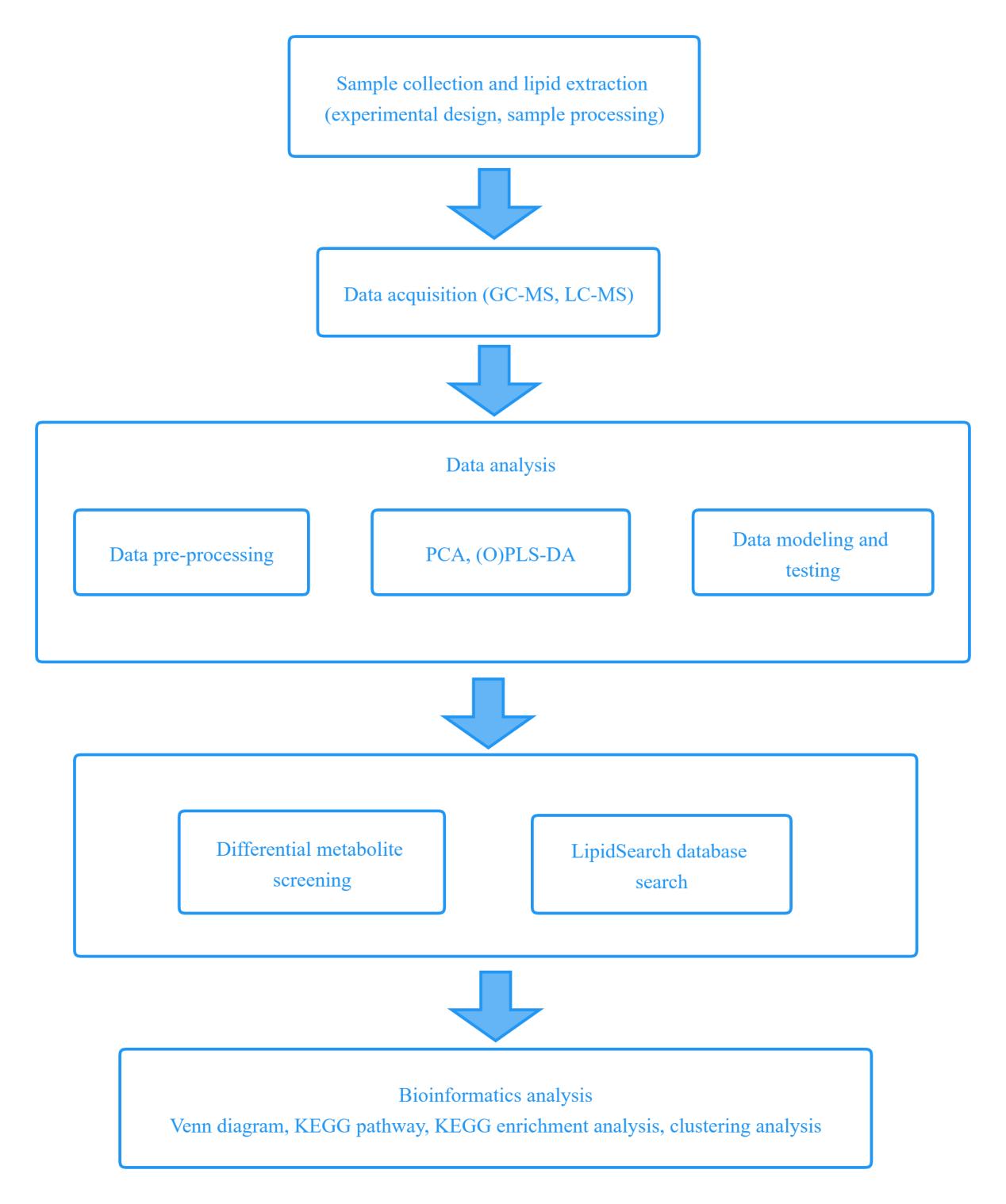
Advantages of our Lipidomics Service
- Advanced platform: Creative Proteomics has LC-MS and GC-MS platforms. Ultra-high resolution, ultra-high sensitivity, fast scanning speed and high-throughput mass spectrometry analysis.
- Professional bioinformatics analysis: We have a professional bioinformatics analysis team for data analysis. Personalized information analysis can be performed according to customer needs.
- Strict quality control: From sample processing, quality evaluation to machine testing, every step is strictly controlled to ensure the accuracy of the results.
- Rich experience in sample pre-processing: completed projects covering plasma, serum, urine, tissue, intestinal contents, feces, saliva, culture fluid, microorganisms, plant samples, etc.
- Excellent qualitative and quantitative capabilities: independent construction of a database of lipid metabolites, including 34 categories, more than 70,000 kinds of lipid substances, more than 100,000 MS/MS secondary mass spectra to qualify and quantify thousands of lipids, with high reproducibility of test results to ensure the accuracy of detection.
- Professional detection and analysis capabilities: strict quality control system, together with ultra-high resolution liquid chromatography-mass spectrometry system and professional data pre-processing and analysis capabilities, providing one-stop solutions for data pre-processing, metabolite identification, statistical analysis, metabolic pathway analysis, etc.
Applications
- Identification of lipid substances in tissue cells.
- Research on lipid function and metabolic regulation.
- Lipid metabolic pathways and network research.
- Lipid biomarker discovery.
Sample Requirements
- Serum and plasma: 300ul.
- Cells: 1×107.
- Cell culture supernatant: 10 ml.
- Feces: 200mg.
- Plant tissues: 500mg.
- Animal tissues: 200mg.
Our experts have many years of experience in lipidomics, bioinformatics, and various application fields from food, agriculture, disease research to pharmacy, can provide you with detailed plans, implementation and provide you with corresponding experiments report. Whether you need a solution for lipidomics or conducting targeted or non-targeted lipid research, we will work closely with you to determine the research purpose and develop a customized plan. If you have any questions or specific requirements, please feel free to contact us.

Reference
- Hyötyläinen T, Orešič M. Analytical lipidomics in metabolic and clinical research. Trends in Endocrinology & Metabolism, 2015, 26(12): 671-673.
- Li Q, Liang X, Xue X, et al. Lipidomics Provides Novel Insights into Understanding the Bee Pollen Lipids Transepithelial Transport and Metabolism in Human Intestinal Cells. Journal of Agricultural and Food Chemistry, 2019, 68(3): 907-917.
Frequently Asked Questions and Precautions:
Is fatty acid testing referring to free fatty acids in biological samples?
No, it is not. Fatty acids in biological samples are primarily present in forms such as triglycerides and phospholipids, and the relative content of free fatty acids is relatively low. In this method, all lipids are hydrolyzed into fatty acids and then methylated (esterified) simultaneously during hydrolysis and methylation. Therefore, the measurement reflects the total fatty acid content of all lipids. However, since gas chromatography measures fatty acid methyl esters, the source of the fatty acids cannot be determined with this method.
Should fatty acid composition be measured using dried or fresh samples?
It is recommended to use dried samples. Firstly, dried samples are convenient for transportation. Secondly, the methylating reagent used in the sample pretreatment is a methanol-sulfuric acid solution. When the sample contains a large amount of water, it can interfere with the methylation process.
Regarding quantification:
The gas chromatograph is coupled with a flame ionization detector (FID). The main characteristic of the FID is its response to almost all volatile organic compounds. The relative response of all hydrocarbon compounds (carbon number ≥ 3) is almost equal, as well as the relative response of isomers of hydrocarbons with heteroatoms (carbon number ≥ 3). This means that during quantification, the ratio of the mass concentration of fatty acid compounds to the internal standard is equal to the ratio of their peak areas.
The mass spectrometer, as a detector, detects ion signals, and the ionization efficiency of compounds varies.
Case: Integrated-Metabolomics and Lipidomics Analysis Reveal Remodeling of Lipid Metabolism and Amino Acid Metabolism in Glucagon Receptor-Deficient Zebrafish
Journal: Front Cell Dev Biol.
Published: 2022
Research Background:
The glucagon receptor (GCGR), activated by glucagon, is essential for glucose, amino acid, and lipid metabolism in animals. Blocking GCGR has been shown to induce various issues, including hypoglycemia, in organisms. However, the mechanisms by which GCGR regulates these physiological functions are not fully understood.
In this study, untargeted metabolomics and lipidomics analyses were performed on both wild-type and GCGR mutant (gcgr−/−) zebrafish to investigate how GCGR regulates these physiological functions.
Research Strategy:

Research Results:
1 Knockout of GCGR induced metabolic disruptions in zebrafish.
Metabolites altered in the gcgr-/- mutant were subjected to KEGG pathway analysis and network analysis, revealing that pathways and substances related to lipid metabolism were the most significantly affected.
 Figure 1: A - Histogram showing the number of pathway types and metabolites; B - Histogram depicting the number of pathways and substances; C - Network analysis.
Figure 1: A - Histogram showing the number of pathway types and metabolites; B - Histogram depicting the number of pathways and substances; C - Network analysis.
2 Remodeling of lipid metabolism and amino acid metabolism in gcgr−/− mutant zebrafish.
Among these altered metabolic pathways, significant changes were observed in pathways related to lipid metabolism, as follows: (1) Significant alterations in saturated fatty acids and unsaturated fatty acids (Figure 2 and Figure 3). (2) Reprogramming of cholesterol metabolism, evident in the pathways of steroid hormone biosynthesis and bile acid biosynthesis (Figure 4). (3) Notable impact on tryptophan metabolism, with all eight altered metabolites in the pathway decreased (Figure 5). (4) Downregulation of urea cycle involved in amino acid catabolism in gcgr−/− mutants (Figure 6).
 Figure 2: GCGR knockout induced dysregulation of glycerophospholipid metabolism.
Figure 2: GCGR knockout induced dysregulation of glycerophospholipid metabolism.
 Figure 3: GCGR knockout affects arachidonic acid metabolism.
Figure 3: GCGR knockout affects arachidonic acid metabolism.
 Figure 4: GCGR gene knockout affects cholesterol metabolism.
Figure 4: GCGR gene knockout affects cholesterol metabolism.
 Figure 5: GCGR knockout affects tryptophan metabolism.
Figure 5: GCGR knockout affects tryptophan metabolism.
 Figure 6: GCGR knockout affects urea production.
Figure 6: GCGR knockout affects urea production.
Creative Proteomics can provide an LC-MS detection platform for untargeted metabolomics, lipidomics, targeted metabolomics, and comprehensive metabolomics solutions. Here, you can find the perfect solution for your research needs, offering worry-free technical services.