- Service Details
- Case Study
- FAQ
What Is Peptidomics
Peptidomics is defined as a systematic, comprehensive, qualitative/quantitative multiplex analysis of endogenous peptides (up to ~20 kDa) in biological samples collected at a chosen time point and location. Peptidomics served as a complement strategy to bridge the gap between proteomics and metabolomics. The invaluable information can be obtained from studies of the numerous soluble polypeptides, including signaling molecules (such as cytokines, growth factors, and peptide hormones), and other small protein fragments and peptides of undetermined functions likely from disease-specific proteolytic cleavage. Peptidomics, serving as a "bridge" between proteomics and metabolomics, sheds light on protein synthesis, processing, degradation, and their regulation under various physiological and pathological conditions. Additionally, it influences gene expression, substance metabolism, and other processes. Hence, peptidomics is regarded as a crucial link connecting proteomics and metabolomics.
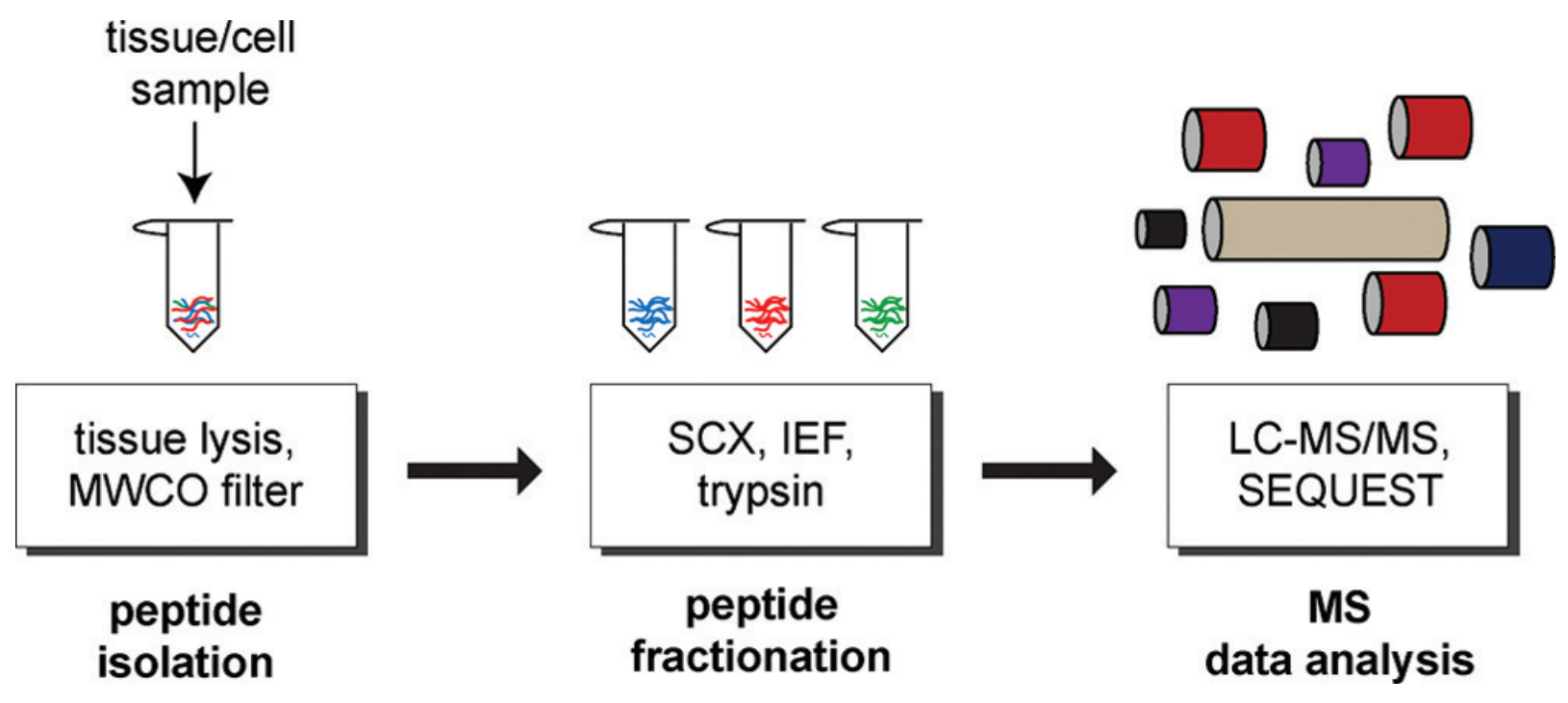 Figure 1. The process of peptidomics analysis (Tinoco, A. D. et al; 2011)
Figure 1. The process of peptidomics analysis (Tinoco, A. D. et al; 2011)
The Significance of Peptidomics Research
Peptidomics enable to decipher a wealth of amino acid sequences information regarding to disease states, drug efficacy or toxicity. Currently, peptidomics sample preparation needs to remove hyper-abundant proteins prior to analysis. Owing to their wide range of functions, it presented unrivaled sensitivity in biomarker discovery and such endogenous peptides possessed great potential as drugs, pharmaceutical agents and biomarker candidates.
In the ongoing quest for health-enhancing compounds that mitigate chronic degenerative diseases and maintain well-being, bioactive peptides represent a class of molecules of great relevance. These peptides naturally occur in food or can be generated through chemical or enzymatic hydrolysis of parent proteins. Participating in a myriad of physiological functions within the human body, peptides contribute to processes spanning gastrointestinal, cardiovascular, immune, endocrine, and nervous systems.
Numerous studies establish a correlation between the consumption of bioactive peptides found in various animal and plant sources and a reduced risk of chronic diseases. Particularly noteworthy are milk and dairy products, considered primary sources of bioactive peptides capable of modulating physiological and metabolic functions, thus exerting a beneficial impact on human health. In comparison to proteins, peptides exhibit a higher bioavailability of essential (indispensable) amino acids and are capable of fulfilling multiple roles that promote health, including mineral transport, immune modulation, antihypertensive effects, antimicrobial activity, and antioxidant properties.
Peptides, as naturally occurring bioactive substances within organisms, have been identified in over ten thousand varieties and serve as signaling molecules, such as hormones, neurotransmitters, and growth factors, contributing to numerous physiological functions. With over seven decades of utilization as therapeutic agents, peptide-based drugs encompass peptides or their modifications for disease prevention, diagnosis, and treatment. An increasing number of peptide-based drugs are developed and clinically applied, effectively addressing conditions like cancer, hepatitis, and diabetes.
Peptide-based drugs primarily originate from endogenous peptides or other exogenous sources. Endogenous peptides are intrinsic to the human body and include substances like enkephalins, thymosins, and pancreatic peptides. Exogenous peptides encompass venom peptides, lectins, antimicrobial peptides from snake, saliva, bee, frog, scorpion, leech, and fly secretions. With the advancement of modern biotechnology, computer-assisted molecular design, screening, and the artificial synthesis of select active peptides are achievable.
Peptide drug discovery strategies involve the identification of extensive peptide sequences, synthesis or recombinant techniques to obtain peptides, followed by target screening for therapeutic applications.
 Nat Rev Drug Discov 2021 04;20(4)
Nat Rev Drug Discov 2021 04;20(4)
Our Advantages
Efficiency: Gentle sample extraction preserves the natural state of peptides, ensuring their authenticity.
High Sensitivity: Leveraging high-resolution mass spectrometry, it offers excellent sensitivity and reproducibility.
Versatility: It enables the identification of both known and unknown peptide sequences, facilitating the discovery of novel active peptides.
Effectiveness: Enhancing the efficiency of endogenous peptide extraction from samples.
High Throughput: Simultaneously studying endogenous peptides across multiple samples.
High Specificity: Independent of antibodies, minimizing cross-reactivity concerns.
Qualitative and Quantitative: Simultaneously performing sequence identification and quantitative analysis of multiple peptides.
Efficient enrichment efficiency, more than 99% of the protein is removed during sample processing.
Comprehensive service capability, simultaneously providing reliable peptide and proteomic data with corresponding bioinformatics analysis.
Peptidomics Analysis at Creative Proteomics
Mass spectrometry and bioinformatic analysis make the proteomics and peptidomics analysis possible. The detected peptides are identified via de novo sequencing, or database searching by tandem mass spectrometers.
Peptides are widely involved in biological process which turns to be ideal biomarker candidates. Due to peptides are available to migrate between different regions of the organism, many pathological processes are supposed to be related with the peptides composition alternation in different body fluids. Changes in peptide abundance have been found to be associated with a variety of diseases. For the screening and identification of peptides, Creative Proteomics can conduct the qualitative and quantitative polypeptides analysis for peptide screening and indentification, for example,amino acid composition, amino acid sequence and molecular weight determination by HPLC, capillary electrophoresis and mass spectrometry.
Comprehensive peptidomics aims to identify and validate all endogenous polypeptides in tested biological samples, In comparison to analysis of expression levels of the target polypeptide in a particular biochemical process. Creative Proteomics can provide comprehensive experiments on peptides with high accuracy and reproducibility.
Our Advantages
- We can provide full steps from separation and enrichment of endogenous peptides to peptide identification and subsequent data analysis.
- Accurate MS based identification of peptide sequences.
- Efficient enrichment efficiency, more than 99% of the protein is removed during sample processing.
- Comprehensive service capability, simultaneously providing reliable peptide and proteomic data with corresponding bioinformatics analysis.
Application Fields
Our peptidomics analysis can be applied to various fields, including but not limited to:
- Biomarker discovery/verification: identification and validation of potential disease biomarkers or drug targets.
- Side effects detection: detection of non-specific and unwanted drug effects, such as degradation.
- Sample quality control: peptide as a degradation product can indicate the quality of the sample.
Based on the superior resources of technology, equipments and personnels, Creative Proteomics offers professional peptide analysis and identification methods for our customers. If you have any questions or specific requirements, please feel free to contact us. We will fulfill your goal.
Peptidome analysis of cerebrospinal fuid in neonates with hypoxic-ischemic brain
Journal: Molecular Brain
Impact Factor: 4.686
Research Background:
Hypoxic-ischemic brain damage (HIBD) can lead to neonatal mortality and severe neurological dysfunction. Current clinical interventions for prevention and treatment are not ideal. Therefore, in this study, the authors identified differentially expressed peptides in cerebrospinal fluid (CSF) from neonates with HIBD and the control group. This identification could provide insights for the diagnosis and treatment of neonatal HIBD, laying the foundation for the development of new therapies.
Sample Strategy:
CSF samples were collected from neonates with HIBD (n = 4) and the control group (n = 4).
Technological Approach:
ITRAQ-labeled peptidomics
Validation Strategy:
Western blot, cell experiments, quantitative fluorescent PCR
Results:
A total of 35 differentially expressed peptides were identified from 25 precursor proteins. In comparison to the control group, the HIBD group exhibited upregulation of one peptide and downregulation of 34 peptides. The authors then investigated potential functional changes in CSF in HIBD by analyzing the cleavage sites of differentially expressed peptides. These cleavage sites displayed regular patterns, allowing for the analysis of proteolytic enzyme functions (as shown in Figure 1). Arginine (R) and lysine (K) were the most common cleavage sites at the N-terminal and C-terminal amino acids of precursor peptides and identified peptides, respectively. Glycine (G) was the most common cleavage site at the C-terminal amino acid of precursor peptides. Lysine (K) and isoleucine (I) were the most common cleavage sites at the N-terminal amino acid of identified peptides.
 Figure 1: Cleavage Sites in Differentially Expressed Peptides
Figure 1: Cleavage Sites in Differentially Expressed Peptides
In order to preliminarily explore the potential functions of these differentially expressed peptides in neonatal HIBD, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted based on precursor proteins. The cellular components of peptide precursors were primarily associated with nuclear chromatin, nucleosome, and chromosomal parts. The most relevant biological processes included chromatin remodeling, chromatin modification, and ATP-dependent chromatin remodeling. The molecular functions of peptide precursors mainly involved chromatin binding, chromatin DNA binding, structure-specific DNA binding, and DNA binding. Pathway analysis revealed that the most relevant pathways associated with these peptide precursors were hypertrophic obstructive cardiomyopathy (HCM), leukocyte transendothelial migration, gastric acid secretion, and arrhythmogenic right ventricular cardiomyopathy (ARVC).
 Figure 2: GO and KEGG Analysis of Peptide Precursors
Figure 2: GO and KEGG Analysis of Peptide Precursors
In order to identify key peptides playing a crucial role in HIBD, the functions of differentially expressed peptides and their precursor proteins were investigated using the STRING database and UniProt database. The protein-protein interaction (PPI) network of precursor proteins is shown in Figure 3. The interactions among these precursor proteins were studied, revealing that heat shock protein 90-alpha (HSP90α/HSP90AA1) occupies a central position in the network and exhibits intricate relationships with other proteins. One of the downregulated peptides in neonatal HIBD is the 2671.5 Da peptide (HSQFIGYPITLFVEKER), which is a fragment of heat shock protein 90-alpha (HSP90α/HSP90AA1). It has been named Hypoxic-Ischemic Brain Damage-Associated Peptide (HIBDAP). Through cellular experiments, the authors speculate that HIBDAP may counteract protective factors in neonatal HIBD through cellular apoptotic mechanisms.
 Figure 3: Protein-Protein Interaction Network of Peptide Precursors
Figure 3: Protein-Protein Interaction Network of Peptide Precursors
This study utilized LC–MS/MS to identify differentially expressed peptides between neonatal HIBD and control group CSF samples, resulting in the identification of 35 differentially expressed peptides. This is the first study to demonstrate differentially expressed peptides in CSF between neonatal HIBD and controls. Furthermore, several significant peptides, including HIBDAP, were discovered, suggesting potential key roles in neonatal HIBD. The molecular mechanisms and biological functions of these peptides may offer new insights into the pathogenesis and therapeutic targets for neonatal HIBD.
References
- Tinoco, A. D.; et al. Investigating endogenous peptides and peptidases using peptidomics. Biochemistry, 2011, 50(35): 7447.
- Hou Xuewen, Yuan Zijun, Wang Xuan et al.PEptidome analysis of cerebrospinal fluid in neonates with hypoxic-ischemic brain damage. Mol Brain, 2020,13:133.
- Muttenthaler, M., King, G.F., Adams, D.J. et al. Trends in peptide drug discovery. Nat Rev Drug Discov 20, 309–325 (2021).
Q: Can peptides be quantitatively detected?
A: Yes, quantitative detection can be achieved through peptidomics. Peptidomics encompasses the comprehensive study of endogenous peptides in biological tissues or fluids. It investigates the structure, function, variations, and associated relationships of endogenous peptides and low-molecular-weight proteins within the organism. The distinction between peptides and proteins is not always clear-cut. Generally, proteins with a molecular weight below 10 kDa are considered peptides. In peptidomics, detection typically focuses on peptide sequences containing six or more amino acids.
Q: How does the experimental workflow of peptidomics differ from conventional proteomics?
A: In peptidomics, there is a distinct step after sample pre-processing involving the separation and enrichment of peptides. Unlike conventional proteomics, peptidomics does not incorporate a quality control process.
Q: Can small peptides with 2-4 amino acids be detected using peptidomics? What is the range of peptide lengths that peptidomics can detect?
A: No, small peptides with 2-4 amino acids cannot be detected using peptidomics. Peptidomics typically focuses on peptides with a length of 6 amino acids or more (commonly in the range of 6-80 amino acids). Peptidomics defaults to the detection of peptides with a molecular weight smaller than 10 kDa.





