Metabolomics can reflect static metabolite abundance, however, the increases in a single metabolite may be due to activation of synthetic pathways or inhibition of the consumption pathways. Therefore, metabolomics is often insufficient to solve all the issues in the study of specific metabolic pathways and metabolic networks. As a complementary tool to metabolomics, metabolic flux analysis is a powerful tool capable of quantifying energy flow (i.e., ATP, cofactor production and consumption) and their balance. Flux variations for a particular metabolic pathway are not necessarily directly related to metabolite concentrations measured at specific time points within that pathway. However, metabolic flux and metabolic pathway kinetics can be defined via targeting mass spectrometry measurements of substrates and isotopes at continuous time points. In addition to the stable isotope 13C, Creative Proteomics also supports the use of 15N to label specific molecules and track their metabolic processes in the organism to obtain information on the dynamics of metabolites in metabolic pathways.
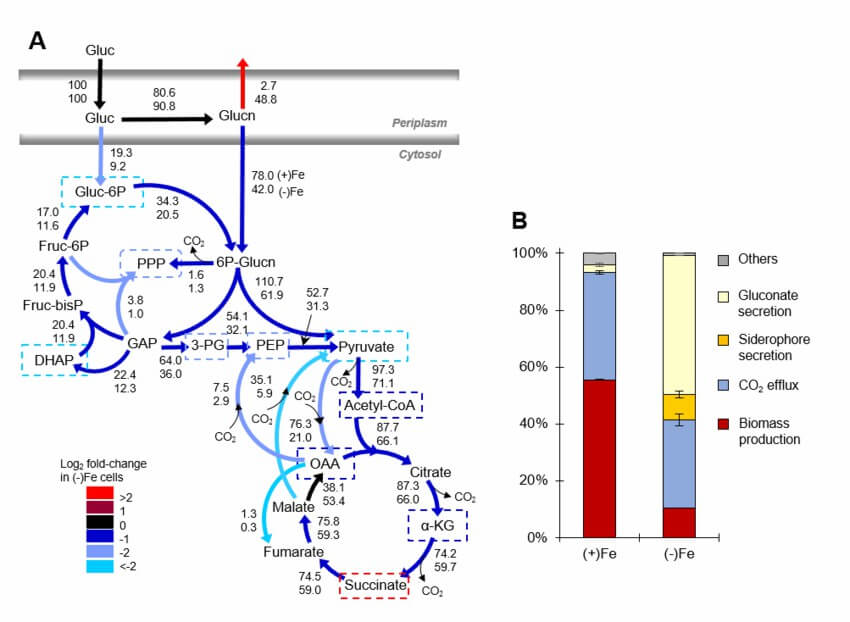 Figure 1. Summary of current 13C-MFA studies on different organisms. (Sasnow, S. S.; et al. 2015)
Figure 1. Summary of current 13C-MFA studies on different organisms. (Sasnow, S. S.; et al. 2015)
Our Services
Creative Proteomics has developed a novel metabolic flux analysis platform to provide 15N-MFA service in a competitive fashion. We can offer a wide range of services to support all research and development activities.
15N-MFA Work Flow
- Experimental Design
Our professional teams can design experiments tailed to meet your study objects and purposes
- 15N Isotopic Labeling and Cell Culture
- Sample Collection and Extraction
Creative Proteomics supports a diversity of samples such as cells, medium, etc
- Sample analysis
We can provide general trend analysis of metabolic group fluxes such as 15N-labeled glutamine for 15N-MFA
- Data analysis
We provide raw data processing by employing mass spectrometry, throughput analysis using proprietary software to quantify throughput and interpret the results
Application of Our Services
- Deep understanding of the metabolic pathways and markers of early diagnosis by comparing the distribution and changes of different metabolic pathways under different environmental conditions, and different metabolic pathways in various metabolic diseases
- Tracing the changes of intermediate metabolites inside and outside the cells, thus identifying the key metabolic pathways and activities of genetically engineered bacteria, and providing a direct basis for improving the synthesis of target metabolites
- Analysis of changes in metabolic functions of cells, tissues and their blood samples and urine before and after genetic modification
Features of Our MFA Platform
- Developed based on the most updated knowledge of biology, bioinformatics and software development
- Widely applicable to a wide range of metabolic system
- Professional bioinformatics teams & personalized bioinformatics analysis services.
- Advanced instrument platform
- Integrated quantitative methodologies and comprehensive solutions for metabolomics
Based on high-performance quantitative techniques and advanced equipment, Creative Proteomics has constantly updating our metabolic flux analysis platform and is committed to offering professional, rapid and high-quality services of 15N-MFA at competitive prices for global customers. Our personalized and comprehensive services can satisfy any innovative scientific study demands, please contact our specialists to discuss your specific needs. We are looking forward to cooperating with you!
Reference
- Sasnow, S. S.; et al. Bypasses in intracellular glucose metabolism in iron-limited Pseudomonas putida. MicrobiologyOpen. 2015.