In structural biology, protein secondary structure is the general three-dimensional form of local segments of proteins. Alpha helices and beta sheets are the most common protein secondary structures. Secondary structure does not describe the specific identity of protein amino acids which are defined as the primary structure, nor the global atomic positions in three-dimensional space, which are recognized to be tertiary structure. Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. Predicting protein tertiary structure from only its amino acid sequence is a very challenging problem, but using the simpler secondary structure definitions is more tractable. Secondary structure prediction is a series of techniques in bioinformatics that aim to predict the secondary structures of proteins sequences based only on knowledge of their primary structure. Now, bioinformaticians at Creative Proteomics are proud to tell you we are open to help you with Protein Secondary Structure Prediction Service!
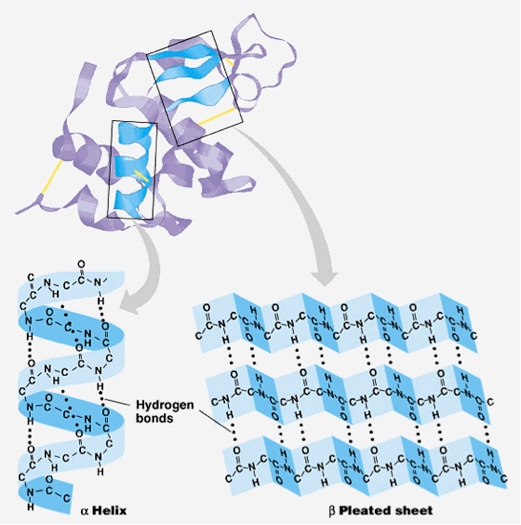
Early methods of secondary-structure prediction were limited to predicting the three predominate states: helix, sheet, and random coil. These methods were generally ~60% accurate in predicting which of the three states amino acid residue adopts. A great increase in accuracy (to nearly ~80%) was made by using multiple sequence alignment; knowing the full distribution of amino acids that occur at a position throughout evolution provides a much better picture of the structural tendencies near that position. For explanation, a given protein might have a glycine at a specific position, which by itself might support a random coil there. Whereas, multiple sequence alignment might indicate that helix-favoring amino acids occur at this position (and nearby positions) in 95% of homologous proteins spanning about a billion years of evolution. What’s more, by examining the average hydrophobicity at that and nearby positions, the same alignment might also suggest a pattern of residue solvent accessibility in compliance with a α-helix. Taken together, these factors would indicate that the glycine of this original protein is α-helical structure, rather than a random coil. Multiple types of methods are applied to combine all the available data to perform a 3-state prediction, such as neural networks, hidden Markov models and support vector machines. Advanced prediction methods can also compute a confidence score for their predictions at every position.
Accuracy of secondary structure prediction is a key element in the prediction of tertiary structure, in all but the simplest (homology modeling) cases. Creative Proteomics can provide variety of secondary structure prediction methods, including:
- Chou & Fasman algorithm
- GOR method
- Neural networks
- Nearest neighbor algorithm
- Hidden Markov models
- Support vector machines
- Combinatorial methods
How to place an order:

*If your organization requires signing of a confidentiality agreement, please contact us by email
As one of the leading omics industry company in the world! Creative Proteomics now is opening to provide secondary structure prediction service for our customers. With rich experience in the field of bioinformatics, we are willing to provide our customer the best secondary structure prediction service! Contact us for all the detailed information!