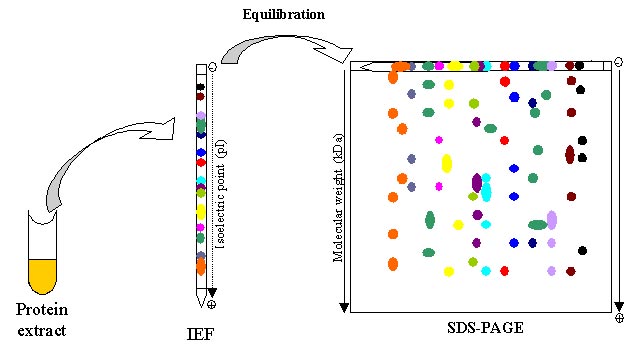
Two-dimensional gel electrophoresis (2-DE) is considered a powerful tool for proteomics work. It is used for separation and fractionation of complex protein mixtures from biological samples. 2-DE separates proteins depending on two different steps: the first one is called isoelectric focusing (IEF) which separates proteins according to isoelectric points (pI); the second step is SDS-polyacrylamide gel electrophoresis (SDS-PAGE) which separates proteins based on the molecular weights(relative molecular weight, Mr). Thus, thousands of proteins can be separated, and the information about IEF and molecular weights can be obtained. 2-DE is a widely used method for protein analysis with the ability to separate thousands of proteins at one time. It can provide direct visual information of changes in protein/post-translational modifications (PTMs) abundance. It can also be used to other analysis, such as whole proteome analysis, detection of biomarkers, drug discovery, and so on. There are several steps for successful 2-DE analysis.
Sample preparation
Speaking of sample preparation, the native samples need to be converted to a physicochemical state suitable for the first dimension IEF and keep the native charge and Mr of the constituent proteins. In order to get good results, appropriate sample preparation is essential. The sample preparation is various because of the difference of the types and origins of proteins. Ideally, the process will result in the complete solubilization, disaggregation, denaturation, and reduction of the proteins in the sample.
First dimension: Isoelectric Focusing (IEF)
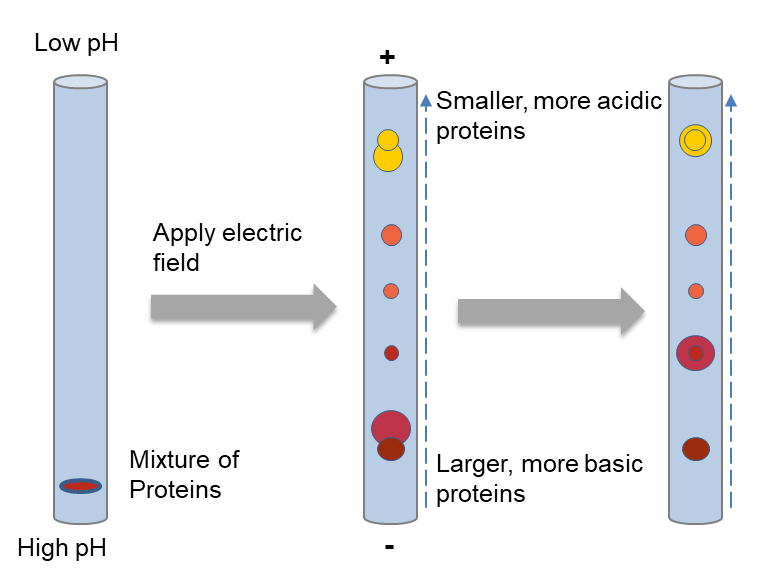
As we mentioned above, the first dimension separate proteins depending on pI of proteins. Proteins are amphoteric molecules and the positive, negative, or zero net charge they carry depending on the pH of the surroundings. The isoelectric point (pI) is defined as the pH of a solution at which the net charge of the protein becomes zero. A protein with a positive net charge will migrate toward the cathode, becoming less positively charged until reaching its pI. While a protein with a negative net charge will migrate toward the anode, becoming less negatively charged until it also reaches its pI.
To be specific, a protein mixture is loaded at the basic end of the pH gradient gel. After applying an electric field, the proteins are separated depending on charges, focusing at positions where the pl value is equivalent to the surrounding pH. Larger proteins will move more slowly through the gel, but with sufficient time will catch up with small proteins of equal charge.
Second dimension: SDS-PAGE
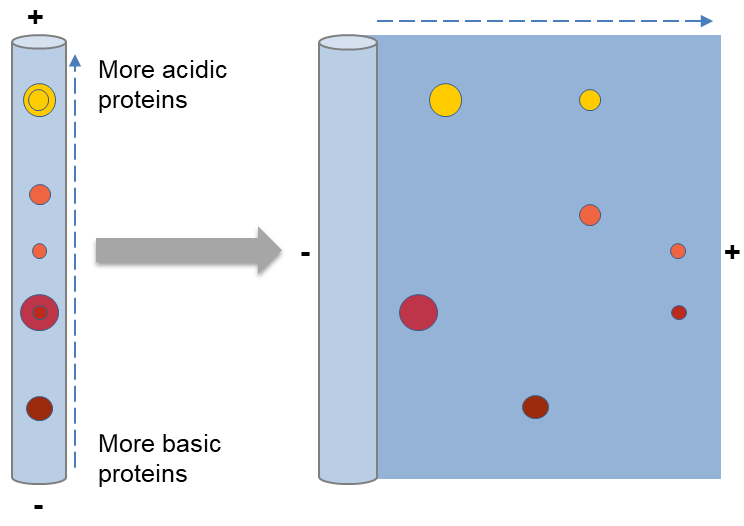
After the first dimension, the second dimension separation can be performed on flatbed or vertical systems on a slab gel. The second dimension is often performed by SDS-PAGE (SDS-polyacrylamide gel electrophoresis), which is an electrophoretic method for separating polypeptides according to their molecular weights (Mr). This method often contains four steps, including preparation of the gel, the equilibrium of the immobilized pH gradient (IPG) strips in SDS buffer, placing the equilibrated IPG strip on the SDS gel, and finally handling the electrophoresis.
SDS can make proteins denaturing and bind to the backbone at a constant molar ratio. When applying SDS and a reducing agent, like a DTT which can cleave disulfide bonds, proteins unfold into linear chains with negative charge proportional to the polypeptide chain length. Polyacrylamide forms a mesh-like matrix which is appropriate for separating proteins. When proteins are separated by SDS-PAGE, smaller proteins migrate faster since the less resistance.
Visualization of results: Staining
There are various methods for visualization of proteins, but the most commonly used are Coomassie Blue staining and silver staining. Silver staining is a sensitive and non-radioactive method. The principle of silver staining is quite simple. The amino acid side chains can bind to silver ions, primary the sulfhydryl and carboxyl groups of proteins, followed by reduction to free metallic silver. As a result, the protein bands are visualized as spots where the reduction occurs. Silver staining is suitable for low protein levels because of its sensitivity (in the very low ng range).
Coomassie Blue staining is a relatively simple method and more quantitative than silver staining. It is suitable to detect protein bands containing about 0.2 μg or more proteins. The Coomassie dye binds to proteins to form a protein-dye complex through Van der Waals attractions. There are two kinds of Coomassie dyes, R250 and G-250.
Further analysis of protein.
There are almost no possibilities to detect the appearance of a few new spots or the disappearance of single spots in large studies with several thousand spots. In addition, evaluation of two gels by manual comparison is also impossible. Therefore, it is necessary to detect differences and obtain information from gels by image collection hardware and image evaluation software. There is some 2D gel analysis software, such as Melanie, PDQuest, Progenesis, REDFIN etc. And the gels can be used for the identification and other applications by mass spectrometry.
References:
- Berkelman T. 2D electrophoresis using immobilized pH gradients, principles and methods. Handbook from Amersham Biosciences, 1998, 57.
- Magdeldin S, Enany S, Yoshida Y, et al. Basics and recent advances of two dimensional-polyacrylamide gel electrophoresis. Clinical proteomics, 2014, 11(1): 16.