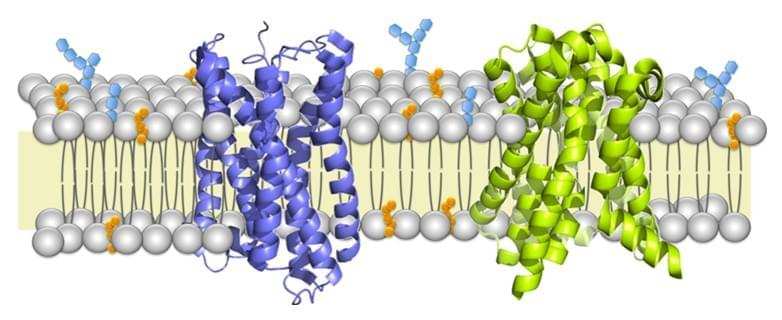
Membrane proteins are a class of proteins that interact with or are part of, biological membranes. Membrane proteins can be classified into three parts based on their location and interactions with membranes: integral (membrane penetrating); peripheral (attached via non-covalent bonds); or lipid-anchored (attached through covalent bonds). Peripheral membrane proteins are attached to one side of the membrane in several ways, like an in-plane a-helix. The integral membrane proteins span across the lipid bilayer and are amphipathic. Their hydrophilic regions protrude into the cytoplasm or the extracellular environment for interaction with soluble proteins and molecules whereas the hydrophobic regions work for the embedding of the proteins into the lipid bilayer. And the lipid-anchored membrane proteins are attached to one side of the membrane through covalent bonds to lipid groups.
Membrane proteins play important roles in various cellular processes, such as cell adhesion, immune response, metabolism and signal transduction. They can act as transporters, receptors and structures proteins. Therefore, membrane proteins are popular targets for proteomics research and the common candidates for drug development. It is reported that about 60% of approved drugs target membrane proteins. However, low abundance, limited solubility, restricted enzyme accessibility are main issues in limiting the amount of information obtained in the study of membrane proteins.
Shotgun proteomics methods have relieved some difficulty in the identification of membrane proteins. However, the major difficulty of membrane proteome analysis still lies in the preparation of membrane proteins. Next, the main processes for membrane protein identification will be identified.
1. Enrichment of membrane protein
Because the integral membrane proteins are low abundant nature, enrichment of membrane protein is essential for proteomic analysis. There are couples of strategies, including: subcellular fractionation (separated by increasing speeds using a glycerol or sorbitol gradient), delipidation (Solubilization of the membrane in the presence of detergent is performed followed by delipidation using a chloroform/methanol solution to extract and solubilize membrane proteins from the lipid bilayers), affinity purification (using biotinylation), and removal of non-membrane proteins (use of high salt and high pH has been successful in removing cytosolic and membrane-associated proteins).
2. Membrane Protein Separation
The membrane protein separation is often divided into gel-based separation and solution-based separation. An earlier gel-based separation is SDS-PAGE prior to mass spectrometry, which is now known as GeLC-MS/MS. In this approach, the gel is cut into slices and digested with trypsin. And the peptides were extracted from gel slices and analyzed by MS/MS. At last, the peptides are identified with databases. There are some other gel-based separation methods, such as blue-native electrophoresis (BNE), clear-native electrophoresis (CNE), and high-resolution clear-native electrophoresis (hrCNE).
Speaking to solution-based separation, multidimensional protein identification technology (MuPIT) is a 2D chromatographic approach to separating proteins, in which proteins are digested into peptides and then the peptides are separated by using strong cation exchange(SCX) and reverse phase chromatography. It is reported that another approach to separating peptides or proteins is immobilized pH gradient isoelectric focusing (IPG-IEF). In this method, digested membrane proteins can be separated by IPG-IEF in the presence of 60% methanol.
3. Protease Digestion
For effective digestion, the backbone of the proteins must be accessible for the proteolytic enzymes. However, access to certain parts of membrane proteins is often blocked by sugars or lipids. Trypsin digestion is a typical way to digestion. However, some membrane proteins tend to lack trypsin cleavage sites, and it leads to large peptide fragments which keep up their hydrophobic nature and are not detected by the mass spectrometer. Proteinase K which is a non-specific protease can be used in membrane proteins. But peptides generated by non-specific proteases are difficult to predict because of the random location of positive charges. Elastase has been well characterized for the analysis of membrane proteins.
4. Mass spectrometry and computational analysis
Tandem mass spectrometry analysis equipped with a MALDI or ESI ion source is often used for membrane protein identification. Peptides were analyzed by mass spectrometry and obtain mass spectra. Subsequently, the bioinformatics tools and software (such as Mascot) are used for membrane protein identification.
Membrane protein identification provides valuable insights into the regulation of their biological processes. Advanced instrumentation that performs higher mass accuracy will allow for higher proteome coverage, which will accelerate the analysis of membrane proteins.
References:
- Lodish H. Molecular cell biology[M]. Macmillan, 2008.
- Gilmore J M, Washburn M P. Advances in shotgun proteomics and the analysis of membrane proteomes. Journal of proteomics, 2010, 73(11): 2078-2091.