Proteins, biomolecules or macromolecules, perform a wide range of functions in organisms. Almost all of the cellular processes require that proteins specifically recognize a multitude of different interaction partners. They can carry out their roles by interacting with other molecules, including DNA, RNA, proteins and small molecules. Protein and protein interactions (PPIs), which refers to intentional physical contacts established between two or more proteins as a result of biochemical events and/or electrostatic forces.
Biological Effects of Protein-Protein Interactions
Protein-Protein interactions play important roles in various biological processes, including cell-to-cell interactions, cell cycle progression, signal transduction, and metabolic pathways. Phizicky and Fields have marked the important properties of PPIs, including:
- PPIs can alter the kinetic properties of enzymes, which may lead to subtle changes in substrate binding or allosteric effects.
- PPIs can act as a general mechanism to allow for substrate channeling by moving a substrate between domains or subunits.
- PPIs can create a novel binding site for small effectors molecules.
- PPI can inactivate or suppress a protein.
- PPI can change the specificity of a protein for its substrate by interacting with different binding partners.
- In an upstream or a downstream event, PPIs can work as a regulatory role.
Types of the Protein-Protein Interactions
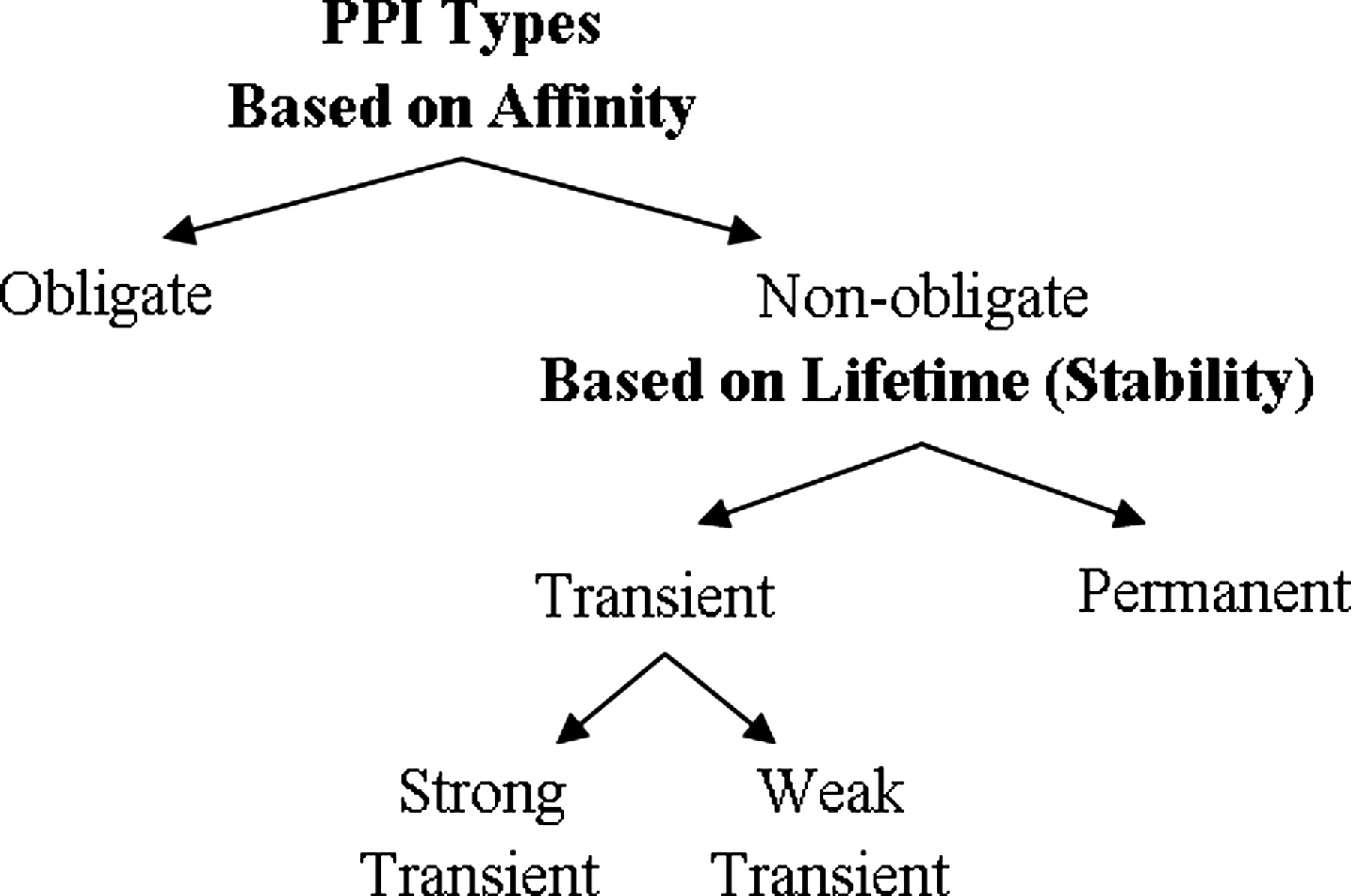
A physical protein interaction can be classified into different interaction types depending on many factors, including composition, affinity, and lifetime.
Based on compositions, these groups of complexes can be classified into homo-oligomeric and hetero-oligomeric complexes. A homo-oligomer is formed when a PPI occurs between identical chains whereas a hetero-oligomer is formed when a PPI occurs between non-identical chains. For example, the cytochrome c’ homodimer is a homodimer (two protein complexes). In addition, GroEL consists of 14 identical subunits of 57 kDa each forming two heptameric rings arranged back-to-back. Several enzymes, carrier proteins, scaffolding proteins, and transcriptional regulatory factors carry out their functions as homo-oligomers.
Obligate and non-obligate complexes are differentiated by the affinity. An obligate interaction means the constituents (protomers, monomers) of a complex are unstable on their own in vivo. The components of non-obligate interactions can exist independently. For instance, Ku proteins, which are involved in DNA repair, are shown to bind DNA as obligate homodimers. H-Ras protein, a G protein, can interchangeably form non-obligate complexes with guanosine triphosphatase (GTPase) activating proteins (GAPs).
According to lifetime, there are transient and permanent interactions. The components of transient interaction associate and dissociate temporarily in vivo while permanent interactions are usually very stable and irreversible. For example, IL-5 cytokine dimer is a permanent protein-protein interaction. In addition, PPIs can be classified into domain-domain and domain-peptide interactions based on folds. The domain-peptide complexes are mostly transient as they are formed by the recognition of a globular domain, a short linear motif (LM) and the small interface on which the interaction takes place.
To summarize, all obligate PPIs are permanent while not all permanent interactions are obligate. Non-obligate interactions are transient but some non-obligate interactions are permanent, like some enzyme−inhibitor interactions. The strong transient category includes protein interactions that shift from an unbound/weakly bound state to a strongly bound state, which is generally triggered by an effector molecule.
Figure 1. Relation of protein-protein interaction types based on affinity and stability. (Acuner Ozbabacan S E, et al., 2011)
Protein and Protein Interactions in Drug Development
Despite the significate roles of PPIs in cellular functions, they have considerable potential roles as therapeutic targets. Targeted modulation of PPIs with small molecules is one of the most promising approaches with more than 50 PPIs that have successfully been targeted with small molecules. Most of these are inhibitors of PPIs, but stabilization of PPIs is also a strategy for targeted PPI modulation.
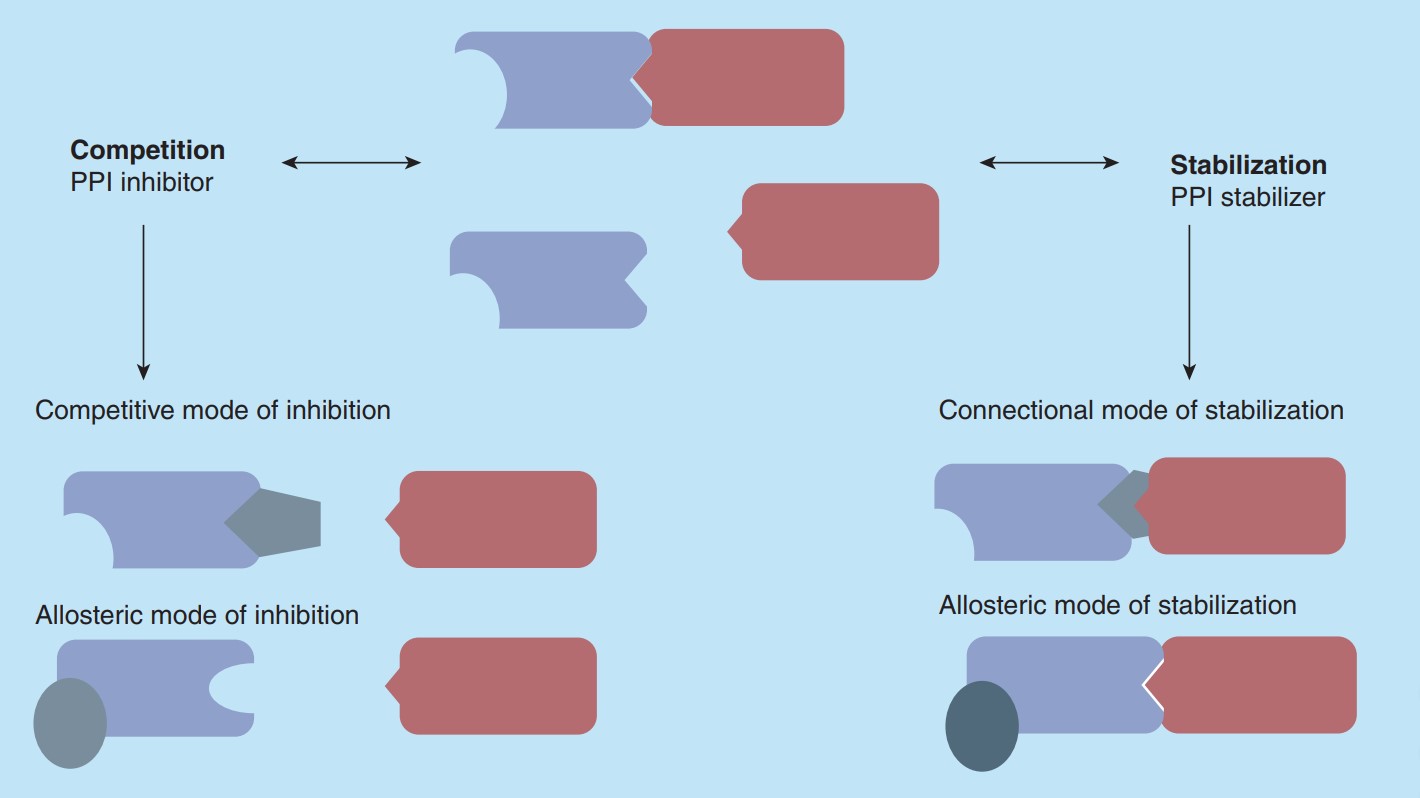
As shown in Figure 2, compounds have different modes of action on PPIs. For PPI inhibitors, they can display a competitive effect on the PPI to prevent the binding of the natural binding partner, which was called competitive mode. Another way is by binding of a compound to a site not-directly part of the PPI interface but arresting the protein in a low-affinity conformation, so that binding of the natural binding partner is impeded. A connectional or allosteric mode of stabilization is also possible. Through binding of a compound in an allosteric mode of stabilization, a protein can bind to its natural ligand in a high-affinity conformation to enhance PPI. Another form of stabilization is the connectional mode of action, which is the direct binding in the rim of the interface with simultaneously contacting both protein partners.
Figure 2. Compound effect on a protein-protein interaction. (Skwarczynska M & Ottmann C, 2015)
There are various methods to detect protein and protein interactions, including yeast two-hybrid screening, affinity purification coupled to mass spectrometry, and so on. If you want to know more about the methods to detect protein-protein interactions, please pay attention to our next blog.
At Creative Proteomics, our team of experts with extensive experience can help you understand what you are trying to investigate and give you the most appropriate solutions about protein and protein interaction. We can provide protein-protein interaction services based on different methods, including:
- Co-immunoprecipitation (co-IP)
- Pull-Down Assay
- Crosslinking Protein Interaction Analysis
- Label Transfer Protein Interaction Analysis
- Far-Western Blot Analysis
References:
- Acuner Ozbabacan S E, Engin H B, Gursoy A, et al. Transient protein-protein interactions. Protein engineering, design and selection, 2011, 24(9): 635-648.
- Rao V S, Srinivas K, Sujini G N, et al. Protein-protein interaction detection: methods and analysis[J]. International journal of proteomics, 2014, 2014.
- Skwarczynska M, Ottmann C. Protein-protein interactions as drug targets. Future medicinal chemistry, 2015, 7(16): 2195-2219.