A precise quantification of individual proteins and global-level proteins can be achieved due to the enhanced sensitivity, resolution, and mass accuracy of MS technology, in conjunction with advancements in HPLC efficiency, software development, and computing power.
MS-based accurate and reliable quantification analysis of alterations in protein abundance and their post-translational modifications (PTMs) between diverse conditions (i.e., healthy versus diseased, normal versus a specific stimulus) provides valuable insights into molecular interactions, signaling pathways, and enables the identification of pathological mechanisms at the phenotypic level as well as biomarkers for improved diagnostics, prediction, and treatment. As a result, quantitative measurements on both protein expression and protein PTMs have been developed and have become an integral part of current proteomic studies.
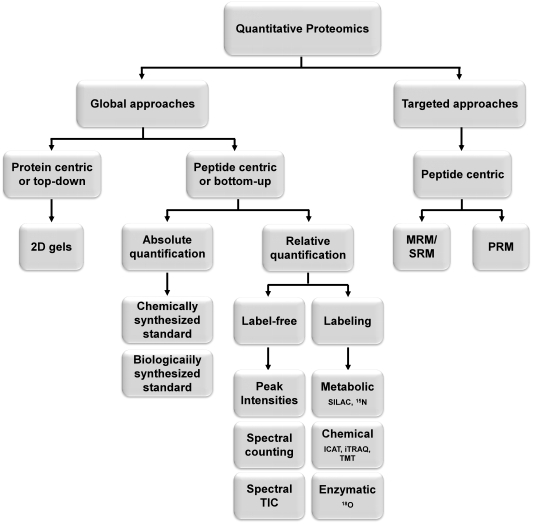 Figure 1. Categorization of the main strategies for quantitative proteomics.
Figure 1. Categorization of the main strategies for quantitative proteomics.
Quantitative Proteomics Strategies
Several technologies and workflows have been presented. According to study aim and scope, two main strategies are used: (i) Unbiased quantification of hundreds or thousands of proteins for the whole protein profile comparison and (ii) targeted quantification of only a few proteins (single or several). Depending on the quantification level, protein-centric or peptide-centric approach could be applied. According to the underlying methodology, MS-based quantification can be further divided into two subgroups: (i) label-based quantification utilizing stable isotope labels incorporated within the peptides/proteins; (ii) label-free quantification in which sample retains its native isotope composition. Depending on the information provided by these quantification methods, they are classified into (i) relative and (ii) absolute. Furthermore, for untargeted quantitative proteomics experiments, the DDA (data-dependent acquisition) or DIA (data independent acquisition) mode can be used. Here are the particular advantages and disadvantages of popular strategies.
Table 1. Overview of the technical parameters of the main strategies and methodologies for quantitative proteomics.
| Quantification method | Description | Number of plex | Quantification level | Advantages | Disadvantages |
|---|---|---|---|---|---|
| ICAT (Isotope-Coded Affinity Tag) | Cysteine-specific chemical labeling on protein/peptide level (in vitro). | 2-plex | MS | 1) Reduced in sample complexity due to affinity enrichment of labeled peptides; 2) Applicable to any sample (cells, animal or human tissue). | 1) Requires cysteine-containing protein/peptides for labeling; 2) Limited by the 2-plex multiplexing capability; 3) Does not allow in vivo labeling. |
| iTRAQ (Isobaric Tags for Relative and Absolute Quantification) | Amine-specific labeling with isobaric tags on the peptide level (in vitro). level. | Up to 8-plex | MS/MS | 1) Efficient labeling enhanced signal intensity in MS and MS/MS, high multiplexing capability, simple data analysis; 2) Applicable to any sample (cells, animal or human tissue); 3) Commercially available. | 1) Expensive; 2) Does not allow in vivo labeling. |
| TMT (Tandem Mass Tag) | Amine-specific labeling with isobaric tags on the peptide level (in vitro). Quantification at level. | Up to 18-plex | MS/MS | 1) Efficient labeling enhanced signal intensity in MS and MS/MS, high multiplexing capability, simple data analysis; 2) Applicable to any sample (cells, animal or human tissue); 3) Commercially available. | 1) Expensive; 2) Requires fragmentation with HCD or ETD; 3) Does not allow in vivo labeling. |
| SILAC (Stable Isotope Labeling by Amino Acids in Cell Culture) | Metabolic labeling with amino acids containing stable heavy isotopes on the protein level (in vivo and ex vivo). | Up to 5-plex | MS | 1) Efficient labeling, one label for (tryptic) peptide, semiautomatic data analysis; 2) Applicable to cells but can be expanded to tissues or model organisms using internal standards (e.g., superSILAC) | 1) High costs, especially when applied to whole organisms; 2) SuperSILAC experiments have reduced quantitative proteome coverage; 3) Requires metabolically active cells to introduce labels. |
| 15N labeling | Metabolic labeling with 15N-enriched media on the Protein level (in vivo and ex vivo). | 2-plex | MS | 1) Efficient labeling. 2) Applicable to cells and model organisms. | 1) Expensive. Complex data analysis; 2) Limited multiplexing capability (up to 2-plex); 3) Not suitable for clinical samples. |
| 18O labeling | Enzymatic labeling with 18O-labeled water on the peptide level (in vitro). | 2-plex | MS | 1) Low costs, simple in handling; 2) Applicable to any sample (cells, animal or human tissue). | 1) Incomplete labeling complicates data analysis; 2) Limited multiplexing capability (up to 2-plex); 3) Not suitable for in vivo labeling. |
| Spectral counting | Relative comparison of different samples based on the number of identified peptides or acquired MS/MS spectra, respectively No labeling required, inexpensive compared to stable isotope labeling methods. | 1) Simplified sample handling, unlimited number of samples; 2) Applicable to any sample (cells, animal or human tissue), | 1) Requires high reproducibility. Longer data acquisition time compared to isotope labeling; 2) Lower accuracy than labeling methods; 3) Requires large sample size (spectral counts) to confidently predict small changes in expression 4) Lower accuracy than labeling and XIC-based LFQ methods. |
The targeted MS approaches, i.e., SRM (selected reaction monitoring), also known as MRM (multiple reaction monitoring), and parallel reaction monitoring (PRM), which are normally not performed using heavy isotope labeling, are highly reproducible and relatively fast. Moreover, SRM and PRM offer its best performance for absolute quantification when applying isotope-labeled peptides/proteins as internal standards.
With years' experience in advanced experiment equipment, Creative Proteomics can provide a variety of proteomics services to assist your scientific research, including:
1) iTRAQ-based proteomics analysis service
2) TMT-based proteomics analysis service
3) SILAC-based proteomics analysis service
4) Absolute quantification (AQUA) service
5) Label-free quantification service
6) Semi-quantitative proteomics analysis service
7) Parallel Reaction Monitoring (PRM)
8) SRM&MRM
As a leading company in the omics industry, Creative Proteomics has been providing customers with a range of protein quantification services for many years. If you're not quite sure which methods are suitable for your research and samples at hands, please feel free to contact us for assistance. Our proteomics experts are glad to help you with your specific proteomics project, with accurate and reliable results and short turnaround time. If you have any questions, Please feel free to contact us and see how we can help to address your problems.
Our customer service representatives are available 24 hours a day, from Monday to Sunday. INQUIRY
References
- Schulze WX, Usadel B. Quantitation in mass-spectrometry-based proteomics. Annual Review of Plant Biology. 2010;61:491-516.
- Rozanova S, Barkovits K, Nikolov M, et al. Quantitative Mass Spectrometry-Based Proteomics: An Overview. Methods in Molecular Biology. 2021;2228:85-116








